A2A Adenosine Receptor Antagonists: Are Triazolotriazine and Purine Scaffolds Interchangeable?
- PMID: 35458588
- PMCID: PMC9032385
- DOI: 10.3390/molecules27082386
A2A Adenosine Receptor Antagonists: Are Triazolotriazine and Purine Scaffolds Interchangeable?
Abstract
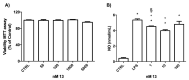
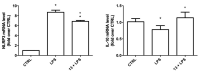
The A2A adenosine receptor (A2AAR) is one of the four subtypes activated by nucleoside adenosine, and the molecules able to selectively counteract its action are attractive tools for neurodegenerative disorders. In order to find novel A2AAR ligands, two series of compounds based on purine and triazolotriazine scaffolds were synthesized and tested at ARs. Compound 13 was also tested in an in vitro model of neuroinflammation. Some compounds were found to possess high affinity for A2AAR, and it was observed that compound 13 exerted anti-inflammatory properties in microglial cells. Molecular modeling studies results were in good agreement with the binding affinity data and underlined that triazolotriazine and purine scaffolds are interchangeable only when 5- and 2-positions of the triazolotriazine moiety (corresponding to the purine 2- and 8-positions) are substituted.
Keywords: A2A adenosine receptor antagonists; anti-Parkinson agents; anti-inflammatory agents; molecular modeling; purine derivatives; triazolotriazine derivatives.
Conflict of interest statement
Authors declare no conflict of interest.
Figures








References
-
- Wilson C.N., Mustafa S.J. Adenosine Receptors in Health and Disease. Volume 193 Springer; Berlin/Heidelberg, Germany: 2009.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

