Reactive Acrylamide-Modified DNA Traps for Accurate Cross-Linking with Cysteine Residues in DNA-Protein Complexes Using Mismatch Repair Protein MutS as a Model
- PMID: 35458636
- PMCID: PMC9031232
- DOI: 10.3390/molecules27082438
Reactive Acrylamide-Modified DNA Traps for Accurate Cross-Linking with Cysteine Residues in DNA-Protein Complexes Using Mismatch Repair Protein MutS as a Model
Abstract
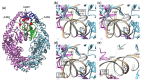
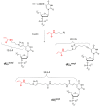
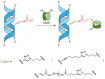
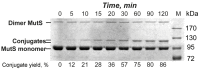
Covalent protein capture (cross-linking) by reactive DNA derivatives makes it possible to investigate structural features by fixing complexes at different stages of DNA-protein recognition. The most common cross-linking methods are based on reactive groups that interact with native or engineered cysteine residues. Nonetheless, high reactivity of most of such groups leads to preferential fixation of early-stage complexes or even non-selective cross-linking. We synthesised a set of DNA reagents carrying an acrylamide group attached to the C5 atom of a 2'-deoxyuridine moiety via various linkers and studied cross-linking with MutS as a model protein. MutS scans DNA for mismatches and damaged nucleobases and can form multiple non-specific complexes with DNA that may cause non-selective cross-linking. By varying the length of the linker between DNA and the acrylamide group and by changing the distance between the reactive nucleotide and a mismatch in the duplex, we showed that cross-linking occurs only if the distance between the acrylamide group and cysteine is optimal within the DNA-protein complex. Thus, acrylamide-modified DNA duplexes are excellent tools for studying DNA-protein interactions because of high selectivity of cysteine trapping.
Keywords: DNA mismatch repair; DNA modification; DNA–protein complex; MutS; crosslinking; modified oligonucleotide; regioselectivity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Hyjek-Składanowska M., Stasińska A.R., Napiórkowska-Gromadzka A., Bartłomiejczak A., Seth P.P., Chmielewski M.K., Nowotny M. Disulfide bridge cross-linking between protein and the RNA backbone as a tool to study RNase H1. Bioorg. Med. Chem. 2020;28:115741. doi: 10.1016/j.bmc.2020.115741. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

