Development and Application of an LC-MS/MS Untargeted Exposomics Method with a Separated Pooled Quality Control Strategy
- PMID: 35458780
- PMCID: PMC9031529
- DOI: 10.3390/molecules27082580
Development and Application of an LC-MS/MS Untargeted Exposomics Method with a Separated Pooled Quality Control Strategy
Abstract
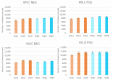
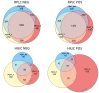
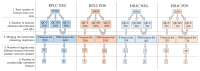
Pooled quality controls (QCs) are usually implemented within untargeted methods to improve the quality of datasets by removing features either not detected or not reproducible. However, this approach can be limiting in exposomics studies conducted on groups of exposed and nonexposed subjects, as compounds present at low levels only in exposed subjects can be diluted and thus not detected in the pooled QC. The aim of this work is to develop and apply an untargeted workflow for human biomonitoring in urine samples, implementing a novel separated approach for preparing pooled quality controls. An LC-MS/MS workflow was developed and applied to a case study of smoking and non-smoking subjects. Three different pooled quality controls were prepared: mixing an aliquot from every sample (QC-T), only from non-smokers (QC-NS), and only from smokers (QC-S). The feature tables were filtered using QC-T (T-feature list), QC-S, and QC-NS, separately. The last two feature lists were merged (SNS-feature list). A higher number of features was obtained with the SNS-feature list than the T-feature list, resulting in identification of a higher number of biologically significant compounds. The separated pooled QC strategy implemented can improve the nontargeted human biomonitoring for groups of exposed and nonexposed subjects.
Keywords: exposomics; liquid chromatography tandem mass spectrometry; pooled quality controls; untargeted metabolomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

