Anti-SARS-CoV-2 potential of Cissampelos pareira L. identified by connectivity map-based analysis and in vitro studies
- PMID: 35459166
- PMCID: PMC9028906
- DOI: 10.1186/s12906-022-03584-3
Anti-SARS-CoV-2 potential of Cissampelos pareira L. identified by connectivity map-based analysis and in vitro studies
Abstract
Background: Viral infections have a history of abrupt and severe eruptions through the years in the form of pandemics. And yet, definitive therapies or preventive measures are not present. Herbal medicines have been a source of various antiviral compounds such as Oseltamivir, extracted using shikimic acid from star anise (Illicium verum) and Acyclovir from Carissa edulis are FDA (Food and Drug Administration) approved antiviral drugs. In this study, we dissect the anti-coronavirus infection activity of Cissampelos pareira L (Cipa) extract using an integrative approach.
Methods: We analysed the signature similarities between predicted antiviral agents and Cipa using the connectivity map ( https://clue.io/ ). Next, we tested the anti-SARS-COV-2 activity of Cipa in vitro. Molecular docking analyses of constituents of with key targets of SARS-CoV2 protein viz. spike protein, RNA‑dependent RNA‑polymerase (RdRp) and 3C‑like proteinase. was also performed. A three-way comparative analysis of Cipa transcriptome, COVID-19 BALF transcriptome and CMAP signatures of small compounds was also performed.
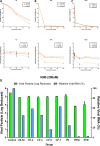
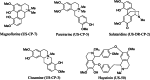
Results: Several predicted antivirals showed a high positive connectivity score with Cipa such as apcidin, emetine, homoharringtonine etc. We also observed 98% inhibition of SARS-COV-2 replication in infected Vero cell cultures with the whole extract. Some of its prominent pure constituents e.g. pareirarine, cissamine, magnoflorine exhibited 40-80% inhibition. Comparison of genes between BALF and Cipa showed an enrichment of biological processes like transcription regulation and response to lipids, to be downregulated in Cipa while being upregulated in COVID-19. CMAP also showed that Triciribine, torin-1 and VU-0365114-2 had positive connectivity with BALF 1 and 2, and negative connectivity with Cipa. Amongst all the tested compounds, Magnoflorine and Salutaridine exhibited the most potent and consistent strong in silico binding profiles with SARS-CoV2 therapeutic targets.
Keywords: Antivirus; BALF; COVID-19; Cissampelos pareira L.; Connectivity map; SARS-CoV-2; Whole plant extract.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of statement.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

