Cisplatin resistance can be curtailed by blunting Bnip3-mediated mitochondrial autophagy
- PMID: 35459212
- PMCID: PMC9033831
- DOI: 10.1038/s41419-022-04741-9
Cisplatin resistance can be curtailed by blunting Bnip3-mediated mitochondrial autophagy
Erratum in
-
Correction: Cisplatin resistance can be curtailed by blunting Bnip3-mediated mitochondrial autophagy.Cell Death Dis. 2022 May 9;13(5):445. doi: 10.1038/s41419-022-04905-7. Cell Death Dis. 2022. PMID: 35534470 Free PMC article. No abstract available.
-
Correction: Cisplatin resistance can be curtailed by blunting Bnip3-mediated mitochondrial autophagy.Cell Death Dis. 2023 Aug 15;14(8):521. doi: 10.1038/s41419-023-05901-1. Cell Death Dis. 2023. PMID: 37582906 Free PMC article. No abstract available.
-
Correction: Cisplatin resistance can be curtailed by blunting Bnip3-mediated mitochondrial autophagy.Cell Death Dis. 2024 Jan 18;15(1):68. doi: 10.1038/s41419-023-06402-x. Cell Death Dis. 2024. PMID: 38238295 Free PMC article. No abstract available.
Abstract
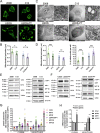
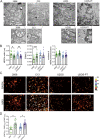
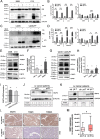
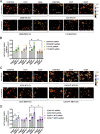
Cisplatin (CDDP) is commonly used to treat a multitude of tumors including sarcomas, ovarian and cervical cancers. Despite recent investigations allowed to improve chemotherapy effectiveness, the molecular mechanisms underlying the development of CDDP resistance remain a major goal in cancer research. Here, we show that mitochondrial morphology and autophagy are altered in different CDDP resistant cancer cell lines. In CDDP resistant osteosarcoma and ovarian carcinoma, mitochondria are fragmented and closely juxtaposed to the endoplasmic reticulum; rates of mitophagy are also increased. Specifically, levels of the mitophagy receptor BNIP3 are higher both in resistant cells and in ovarian cancer patient samples resistant to platinum-based treatments. Genetic BNIP3 silencing or pharmacological inhibition of autophagosome formation re-sensitizes these cells to CDDP. Our study identifies inhibition of BNIP3-driven mitophagy as a potential therapeutic strategy to counteract CDDP resistance in ovarian carcinoma and osteosarcoma.
© 2022. The Author(s).
Conflict of interest statement
ADM is employee and shareholder of Sprint Bioscience AB. All the other authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

