GC/MS-based 13C metabolic flux analysis resolves the parallel and cyclic photomixotrophic metabolism of Synechocystis sp. PCC 6803 and selected deletion mutants including the Entner-Doudoroff and phosphoketolase pathways
- PMID: 35459213
- PMCID: PMC9034593
- DOI: 10.1186/s12934-022-01790-9
GC/MS-based 13C metabolic flux analysis resolves the parallel and cyclic photomixotrophic metabolism of Synechocystis sp. PCC 6803 and selected deletion mutants including the Entner-Doudoroff and phosphoketolase pathways
Abstract
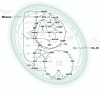
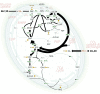
Background: Cyanobacteria receive huge interest as green catalysts. While exploiting energy from sunlight, they co-utilize sugar and CO2. This photomixotrophic mode enables fast growth and high cell densities, opening perspectives for sustainable biomanufacturing. The model cyanobacterium Synechocystis sp. PCC 6803 possesses a complex architecture of glycolytic routes for glucose breakdown that are intertwined with the CO2-fixing Calvin-Benson-Bassham (CBB) cycle. To date, the contribution of these pathways to photomixotrophic metabolism has remained unclear.
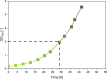
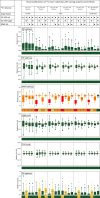
Results: Here, we developed a comprehensive approach for 13C metabolic flux analysis of Synechocystis sp. PCC 6803 during steady state photomixotrophic growth. Under these conditions, the Entner-Doudoroff (ED) and phosphoketolase (PK) pathways were found inactive but the microbe used the phosphoglucoisomerase (PGI) (63.1%) and the oxidative pentose phosphate pathway (OPP) shunts (9.3%) to fuel the CBB cycle. Mutants that lacked the ED pathway, the PK pathway, or phosphofructokinases were not affected in growth under metabolic steady-state. An ED pathway-deficient mutant (Δeda) exhibited an enhanced CBB cycle flux and increased glycogen formation, while the OPP shunt was almost inactive (1.3%). Under fluctuating light, ∆eda showed a growth defect, different to wild type and the other deletion strains.
Conclusions: The developed approach, based on parallel 13C tracer studies with GC-MS analysis of amino acids, sugars, and sugar derivatives, optionally adding NMR data from amino acids, is valuable to study fluxes in photomixotrophic microbes to detail. In photomixotrophic cells, PGI and OPP form glycolytic shunts that merge at switch points and result in synergistic fueling of the CBB cycle for maximized CO2 fixation. However, redirected fluxes in an ED shunt-deficient mutant and the impossibility to delete this shunt in a GAPDH2 knockout mutant, indicate that either minor fluxes (below the resolution limit of 13C flux analysis) might exist that could provide catalytic amounts of regulatory intermediates or alternatively, that EDA possesses additional so far unknown functions. These ideas require further experiments.
Keywords: 13C metabolic flux analysis; CO2; Calvin-Benson-Bassham cycle; Cyanobacteria; Entner-Doudoroff pathway; GC–MS; Glucose; Glycolytic shunt; NMR; Oxidative pentose phosphate pathway; Phosphoketolase pathway; TCA cycle, photomixotrophic growth.
© 2022. The Author(s).
Conflict of interest statement
All authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

