Mechanistic insight of SARS-CoV-2 infection using human hepatobiliary organoids
- PMID: 35459707
- PMCID: PMC9763169
- DOI: 10.1136/gutjnl-2021-326617
Mechanistic insight of SARS-CoV-2 infection using human hepatobiliary organoids
Keywords: HEPATOBILIARY DISEASE.
Conflict of interest statement
Competing interests: None declared.
Figures
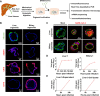
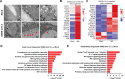
Comment on
-
Liver function test abnormalities at hospital admission are associated with severe course of SARS-CoV-2 infection: a prospective cohort study.Gut. 2021 Oct;70(10):1925-1932. doi: 10.1136/gutjnl-2020-323800. Epub 2021 Jan 29. Gut. 2021. PMID: 33514597 Free PMC article.
References
-
- Chai X, Hu L, Zhang Y. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv 2020.
-
- Gounden V, Vashisht R, Jialal I. Hypoalbuminemia. In: StatPearls. Treasure Island (FL): StatPearls Publishing, 2022. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
