Integrated clinical and genomic evaluation of guadecitabine (SGI-110) in peripheral T-cell lymphoma
- PMID: 35459873
- PMCID: PMC9162925
- DOI: 10.1038/s41375-022-01571-8
Integrated clinical and genomic evaluation of guadecitabine (SGI-110) in peripheral T-cell lymphoma
Abstract
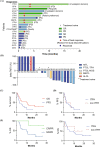
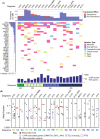
Peripheral T-cell lymphoma (PTCL) is a rare, heterogenous malignancy with dismal outcomes at relapse. Hypomethylating agents (HMA) have an emerging role in PTCL, supported by shared mutations with myelodysplasia (MDS). Response rates to azacitidine in PTCL of follicular helper cell origin are promising. Guadecitabine is a decitabine analogue with efficacy in MDS. In this phase II, single-arm trial, PTCL patients received guadecitabine on days 1-5 of 28-day cycles. Primary end points were overall response rate (ORR) and safety. Translational sub-studies included cell free plasma DNA sequencing and functional genomic screening using an epigenetically-targeted CRISPR/Cas9 library to identify response predictors. Among 20 predominantly relapsed/refractory patients, the ORR was 40% (10% complete responses). Most frequent grade 3-4 adverse events were neutropenia and thrombocytopenia. At 10 months median follow-up, median progression free survival (PFS) and overall survival (OS) were 2.9 and 10.4 months respectively. RHOAG17V mutations associated with improved PFS (median 5.47 vs. 1.35 months; Wilcoxon p = 0.02, Log-Rank p = 0.06). 4/7 patients with TP53 variants responded. Deletion of the histone methyltransferase SETD2 sensitised to HMA but TET2 deletion did not. Guadecitabine conveyed an acceptable ORR and toxicity profile; decitabine analogues may provide a backbone for future combinatorial regimens co-targeting histone methyltransferases.
© 2022. The Author(s).
Conflict of interest statement
JS has received research funding from Astex Pharmaceuticals Inc. related to the published work. All other disclosures for JS and co-authors are outside of the published work. JS has received research funding from Amgen and Bristol Myers Squibb/Celgene; JS has served on Advisory Boards for Novartis, BMS, Mundipharma and Astellas. RG has received honoraria from Merck Sharp & Dohme, Takeda, Novartis, AbbVie and Astellas. EAH has provided consultancy to Specialised Therapeutics; EAH has received research funding from Bristol Myers Squibb/Celgene, Merck KGaA, Astra Zeneca and Roche; EAH has received speakers bureau from Roche, Astra Zeneca, Janssen, Regeneron and Abbvie; EAH has served on Advisory Boards for Roche, Antigene, Bristol Myers Squibb and Astra Zeneca and has received travel expenses from Roche. LMK has received research funding and consultancy fees from Agios Pharmaceuticals and Celgene. JR has received research funding from AbbVie and is a shareholder in Novartis AG and Alcon. GPG has served on Advisory Boards for Roche, Novartis, Janssen and Gilead; GPG has received honoraria/speakers’ bureau from Roche, Novartis, Janssen and Astra Zeneca; GPG has received research funding from Beigene, Merck, Abbvie and Janssen; GPG has received travel funds from Roche and Novartis. SO has provided consultancy to AbbVie, Astra Zeneca, Janssen and Roche; SO has received research funding from Amgen and Beigene; SO has received honoraria from AbbVie, Astra Zeneca, Celgene, CSL Behring, Gilead, Janssen, Merck, Roche and Takeda; SO has served on Advisory Boards for AbbVie, Astra Zeneca, Celgene, CSL Behring, Gilead, Janssen, Merck, Roche and Takeda. The other authors have no COI to disclose.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

