Disentangling Signatures of Selection Before and After European Colonization in Latin Americans
- PMID: 35460423
- PMCID: PMC9034689
- DOI: 10.1093/molbev/msac076
Disentangling Signatures of Selection Before and After European Colonization in Latin Americans
Abstract
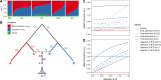
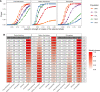
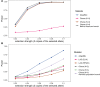
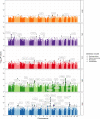
Throughout human evolutionary history, large-scale migrations have led to intermixing (i.e., admixture) between previously separated human groups. Although classical and recent work have shown that studying admixture can yield novel historical insights, the extent to which this process contributed to adaptation remains underexplored. Here, we introduce a novel statistical model, specific to admixed populations, that identifies loci under selection while determining whether the selection likely occurred post-admixture or prior to admixture in one of the ancestral source populations. Through extensive simulations, we show that this method is able to detect selection, even in recently formed admixed populations, and to accurately differentiate between selection occurring in the ancestral or admixed population. We apply this method to genome-wide SNP data of ∼4,000 individuals in five admixed Latin American cohorts from Brazil, Chile, Colombia, Mexico, and Peru. Our approach replicates previous reports of selection in the human leukocyte antigen region that are consistent with selection post-admixture. We also report novel signals of selection in genomic regions spanning 47 genes, reinforcing many of these signals with an alternative, commonly used local-ancestry-inference approach. These signals include several genes involved in immunity, which may reflect responses to endemic pathogens of the Americas and to the challenge of infectious disease brought by European contact. In addition, some of the strongest signals inferred to be under selection in the Native American ancestral groups of modern Latin Americans overlap with genes implicated in energy metabolism phenotypes, plausibly reflecting adaptations to novel dietary sources available in the Americas.
Keywords: Latin Americans; Native Americans; admixture; natural selection.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
-
- Cavalli-Sforza LL editor.; 1969.
-
- Acuña-Alonzo V, Flores-Dorantes T, Kruit JK, Villarreal-Molina T, Arellano-Campos O, Hünemeier T, Moreno-Estrada A, Ortiz-López MG, Villamil-Ramírez H, León-Mimila P, et al. . 2010. A functional ABCA1 gene variant is associated with low HDL-cholesterol levels and shows evidence of positive selection in Native Americans. Hum Mol Genet. 19:2877–2885. - PMC - PubMed
-
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automatic Control 19:716–723.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

