SARS-CoV-2 infection of human-induced pluripotent stem cells-derived lung lineage cells evokes inflammatory and chemosensory responses by targeting mitochondrial pathways
- PMID: 35460571
- PMCID: PMC9088312
- DOI: 10.1002/jcp.30755
SARS-CoV-2 infection of human-induced pluripotent stem cells-derived lung lineage cells evokes inflammatory and chemosensory responses by targeting mitochondrial pathways
Abstract
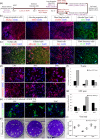
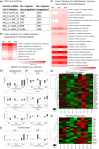
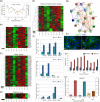
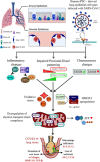
The COVID-19 disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) primarily affects the lung, particularly the proximal airway and distal alveolar cells. NKX2.1+ primordial lung progenitors of the foregut (anterior) endoderm are the developmental precursors to all adult lung epithelial lineages and are postulated to play an important role in viral tropism. Here, we show that SARS-CoV-2 readily infected and replicated in human-induced pluripotent stem cell-derived proximal airway cells, distal alveolar cells, and lung progenitors. In addition to the upregulation of antiviral defense and immune responses, transcriptomics data uncovered a robust epithelial cell-specific response, including perturbation of metabolic processes and disruption in the alveolar maturation program. We also identified spatiotemporal dysregulation of mitochondrial heme oxygenase 1 (HMOX1), which is associated with defense against antioxidant-induced lung injury. Cytokines, such as TNF-α, INF-γ, IL-6, and IL-13, were upregulated in infected cells sparking mitochondrial ROS production and change in electron transport chain complexes. Increased mitochondrial ROS then activated additional proinflammatory cytokines leading to an aberrant cell cycle resulting in apoptosis. Notably, we are the first to report a chemosensory response resulting from SARS-CoV-2 infection similar to that seen in COVID-19 patients. Some of our key findings were validated using COVID-19-affected postmortem lung tissue sections. These results suggest that our in vitro system could serve as a suitable model to investigate the pathogenetic mechanisms of SARS-CoV-2 infection and to discover and test therapeutic drugs against COVID-19 or its consequences.
Keywords: SARS-CoV-2; chemosensory response; induced pluripotent stem cells; inflammatory response; lung epithelial cells; mitochondrial damage.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection.Protein Cell. 2021 Sep;12(9):717-733. doi: 10.1007/s13238-020-00811-w. Epub 2020 Dec 12. Protein Cell. 2021. PMID: 33314005 Free PMC article.
-
Micro-patterned culture of iPSC-derived alveolar and airway cells distinguishes SARS-CoV-2 variants.Stem Cell Reports. 2024 Apr 9;19(4):545-561. doi: 10.1016/j.stemcr.2024.02.011. Epub 2024 Mar 28. Stem Cell Reports. 2024. PMID: 38552631 Free PMC article.
-
A therapy for suppressing canonical and noncanonical SARS-CoV-2 viral entry and an intrinsic intrapulmonary inflammatory response.Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2408109121. doi: 10.1073/pnas.2408109121. Epub 2024 Jul 19. Proc Natl Acad Sci U S A. 2024. PMID: 39028694 Free PMC article.
-
Induced pluripotent stem cell-based disease modeling and prospective immune therapy for coronavirus disease 2019.Cytotherapy. 2022 Mar;24(3):235-248. doi: 10.1016/j.jcyt.2021.08.003. Epub 2021 Sep 14. Cytotherapy. 2022. PMID: 34656419 Free PMC article. Review.
-
SARS-CoV-2 Infection and Disease Modelling Using Stem Cell Technology and Organoids.Int J Mol Sci. 2021 Feb 26;22(5):2356. doi: 10.3390/ijms22052356. Int J Mol Sci. 2021. PMID: 33652988 Free PMC article. Review.
Cited by
-
Mitochondria in COVID-19: from cellular and molecular perspective.Front Physiol. 2024 Jun 21;15:1406635. doi: 10.3389/fphys.2024.1406635. eCollection 2024. Front Physiol. 2024. PMID: 38974521 Free PMC article. Review.
-
The Orf9b protein of SARS-CoV-2 modulates mitochondrial protein biogenesis.J Cell Biol. 2023 Oct 2;222(10):e202303002. doi: 10.1083/jcb.202303002. Epub 2023 Sep 8. J Cell Biol. 2023. PMID: 37682539 Free PMC article.
-
Is There such a Thing as Post-Viral Depression?: Implications for Precision Medicine.Biomol Ther (Seoul). 2024 Nov 1;32(6):659-684. doi: 10.4062/biomolther.2024.170. Epub 2024 Oct 21. Biomol Ther (Seoul). 2024. PMID: 39428555 Free PMC article. Review.
References
-
- Abo, K. M. , Ma, L. , Matte, T. , Huang, J. , Alysandratos, K. D. , Werder, R. B. , Mithal, A. , Beermann, M. L. , Lindstrom‐Vautrin, J. , Mostoslavsky, G. , Ikonomou, L. , Kotton, D. N. , Hawkins, F. , Wilson, A. , & Villacorta‐Martin, C. (2020). Human iPSC‐derived alveolar and airway epithelial cells can be cultured at air‐liquid interface and express SARS‐CoV‐2 host factors. bioRxiv: The Preprint Server for Biology , 132639. 10.1101/2020.06.03.132639 - DOI
-
- Amara, N. , Goven, D. , Prost, F. , Muloway, R. , Crestani, B. , & Boczkowski, J. (2010). NOX4/NADPH oxidase expression is increased in pulmonary fibroblasts from patients with idiopathic pulmonary fibrosis and mediates TGF‐B1‐induced fibroblast differentiation into myofibroblasts. Thorax, 65(8), 733–738. 10.1136/thx.2009.113456 - DOI - PMC - PubMed
-
- Anantharaj, A. , Gujjar, S. , Verma, N. , Khan, N. A. , Shaman, H. , Sharanabasava, P. , Das, A. , Pandey, R. , Pandey, A. K. , & Medigeshi, G. R. (2021). Resolution of viral load in mild COVID‐19 patients is associated with both innate and adaptive immune responses. Journal of Clinical Virology, 21(146), 105060. 10.1016/j.jcv.2021.105060 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

