Circular RNA vaccines against SARS-CoV-2 and emerging variants
- PMID: 35460644
- PMCID: PMC8971115
- DOI: 10.1016/j.cell.2022.03.044
Circular RNA vaccines against SARS-CoV-2 and emerging variants
Abstract
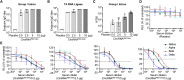
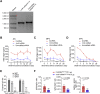
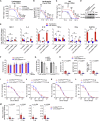
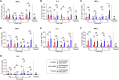
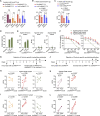
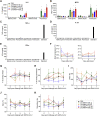
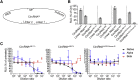
As the emerging variants of SARS-CoV-2 continue to drive the worldwide pandemic, there is a constant demand for vaccines that offer more effective and broad-spectrum protection. Here, we report a circular RNA (circRNA) vaccine that elicited potent neutralizing antibodies and T cell responses by expressing the trimeric RBD of the spike protein, providing robust protection against SARS-CoV-2 in both mice and rhesus macaques. Notably, the circRNA vaccine enabled higher and more durable antigen production than the 1mΨ-modified mRNA vaccine and elicited a higher proportion of neutralizing antibodies and distinct Th1-skewed immune responses. Importantly, we found that the circRNARBD-Omicron vaccine induced effective neutralizing antibodies against the Omicron but not the Delta variant. In contrast, the circRNARBD-Delta vaccine protected against both Delta and Omicron or functioned as a booster after two doses of either native- or Delta-specific vaccination, making it a favorable choice against the current variants of concern (VOCs) of SARS-CoV-2.
Keywords: COVID-19; Delta; Omicron; SARS-CoV-2; circRNA vaccine; circular RNA; mRNA vaccine; vaccine; variant of concern.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Patents related to the data presented have been filed. W.W. is the founder of Therorna, Inc.
Figures





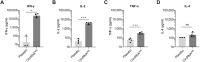


References
-
- Alameh M.G., Tombácz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892.e7. - PMC - PubMed
-
- Boutros C., Tarhini A., Routier E., Lambotte O., Ladurie F.L., Carbonnel F., Izzeddine H., Marabelle A., Champiat S., Berdelou A., et al. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat. Rev. Clin. Oncol. 2016;13:473–486. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

