Omicron-included mutation-induced changes in epitopes of SARS-CoV-2 spike protein and effectiveness assessments of current antibodies
- PMID: 35461370
- PMCID: PMC9034971
- DOI: 10.1186/s43556-022-00074-3
Omicron-included mutation-induced changes in epitopes of SARS-CoV-2 spike protein and effectiveness assessments of current antibodies
Abstract
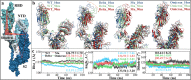
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is spreading globally and continues to rage, posing a serious threat to human health and life quality. Antibody therapy and vaccines both have shown great efficacy in the prevention and treatment of COVID-19, whose development progress and adaptation range have attracted wide attention. However, with the emergence of variant strains of SARS-CoV-2, the neutralization activity of therapeutic or vaccine-induced antibodies may be reduced, requiring long-term virus monitoring and drug upgrade in response to its evolution. In this paper, conformational changes including continuous epitopes (CPs), discontinuous epitopes (DPs) and recognition interfaces of the three representative SARS-CoV-2 spike protein (SP) mutants (i.e., the Delta (B.1.617.2), Mu (B.1.621) and Omicron (B.1.1.529) strains), were analyzed to evaluate the effectiveness of current mainstream antibodies. The results showed that the conformation of SP wild type (WT) and mutants both remained stable, while the local antigenic epitopes underwent significant changes. Sufficient flexibility of SP CPs is critical for effective antibody recognition. The DPs of Delta, Mu and Omicron variants have showed stronger binding to human angiotensin converting enzyme-2 (hACE2) than WT; the possible drug resistance mechanisms of antibodies against three different epitopes (i.e., NTD_DP, RBD1_DP and RBD2_DP) were also proposed, respectively; the RBD2 of Delta, NTD of Mu, NTD and RBD2 of Omicron are deserve more attention in the subsequent design of next-generation vaccines. The simulation results not only revealed structural characteristics of SP antigenic epitopes, but also provided guidance for antibody modification, vaccine design and effectiveness evaluation.
Keywords: Antibody; Drug resistance; Epitopes; SARS-CoV-2; Spike protein.
© 2022. The Author(s).
Conflict of interest statement
No conflict of interest exists in the submission of this manuscript. I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and is not under consideration for publication elsewhere, in whole or in part. All the authors listed have approved the manuscript that is enclosed.
All the authors declare no conflict of interest, financial or otherwise.
Figures




Similar articles
-
A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection.J Virol. 2022 Sep 14;96(17):e0011822. doi: 10.1128/jvi.00118-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972290 Free PMC article.
-
A Bispecific Antibody Targeting RBD and S2 Potently Neutralizes SARS-CoV-2 Omicron and Other Variants of Concern.J Virol. 2022 Aug 24;96(16):e0077522. doi: 10.1128/jvi.00775-22. Epub 2022 Aug 2. J Virol. 2022. PMID: 35916510 Free PMC article.
-
Molecular recognition of SARS-CoV-2 spike protein with three essential partners: exploring possible immune escape mechanisms of viral mutants.J Mol Model. 2023 Mar 24;29(4):109. doi: 10.1007/s00894-023-05509-4. J Mol Model. 2023. PMID: 36964244 Free PMC article.
-
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants.Elife. 2022 Aug 25;11:e78633. doi: 10.7554/eLife.78633. Elife. 2022. PMID: 36004719 Free PMC article.
-
Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.J Virol. 2022 Aug 24;96(16):e0048022. doi: 10.1128/jvi.00480-22. Epub 2022 Aug 4. J Virol. 2022. PMID: 35924918 Free PMC article.
Cited by
-
Is This All COVID-19's Fault? A Study on Trainees in One of the Most Affected Italian Cities.Int J Environ Res Public Health. 2022 Oct 12;19(20):13136. doi: 10.3390/ijerph192013136. Int J Environ Res Public Health. 2022. PMID: 36293715 Free PMC article.
-
Revealing Allosteric Mechanism of Amino Acid Binding Proteins from Open to Closed State.Molecules. 2023 Oct 17;28(20):7139. doi: 10.3390/molecules28207139. Molecules. 2023. PMID: 37894619 Free PMC article.
-
The Relationship between the Transmission of Different SARS-CoV-2 Strains and Air Quality: A Case Study in China.Int J Environ Res Public Health. 2023 Jan 20;20(3):1943. doi: 10.3390/ijerph20031943. Int J Environ Res Public Health. 2023. PMID: 36767307 Free PMC article.
-
A robust, highly multiplexed mass spectrometry assay to identify SARS-CoV-2 variants.medRxiv [Preprint]. 2022 May 29:2022.05.28.22275691. doi: 10.1101/2022.05.28.22275691. medRxiv. 2022. Update in: Microbiol Spectr. 2022 Oct 26;10(5):e0173622. doi: 10.1128/spectrum.01736-22. PMID: 35665019 Free PMC article. Updated. Preprint.
-
Marine sulfated glycans inhibit the interaction of heparin with S-protein of SARS-CoV-2 Omicron XBB variant.Glycoconj J. 2024 Apr;41(2):163-174. doi: 10.1007/s10719-024-10150-1. Epub 2024 Apr 20. Glycoconj J. 2024. PMID: 38642280 Free PMC article.
References
-
- Li Y, Undurraga EA , Zubizarreta J R. Efficacy of localized lockdowns in the SARS-CoV-2 pandemic. medRxiv. 2020;1:1–20. doi: 10.1101/2020.08.25.20182071
-
- Johns Hopkins University. COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE). 2021. https://arcg.is/0fHmTX). Accessed 07 Apr 2021.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
