Mitogenomics and phylogenetics of twelve species of African Saturniidae (Lepidoptera)
- PMID: 35462770
- PMCID: PMC9022641
- DOI: 10.7717/peerj.13275
Mitogenomics and phylogenetics of twelve species of African Saturniidae (Lepidoptera)
Abstract
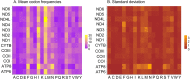
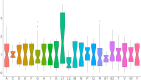
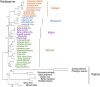
African Saturniidae (Lepidoptera) include numerous species consumed at the caterpillar stage throughout the continent, and their importance to local communities as a source of nutrition and seasonal income cannot be overestimated. However, baseline genetic data with utility for the characterization of their diversity, phylogeography and phylogenetic relationships have remained scarce compared to their Asian counterparts. To bridge this gap, we sequenced the mitochondrial genomes of 12 species found in southern Africa for comparative mitogenomics and phylogenetic reconstruction of the family, including the first representatives of the tribes Eochroini and Micragonini. Mitochondrial gene content and organization were conserved across all Saturniidae included in the analyses. The phylogenetic positions of the 12 species were assessed in the context of publicly available mitogenomes using Bayesian inference and maximum likelihood (ML) methods. The monophyly of the tribes Saturniini, Attacini, Bunaeini and Micragonini, the sister relationship between Saturniini and Attacini, and the placement of Eochroa trimenii and Rhodinia fugax in the tribes Eochroini and Attacini, respectively, were strongly supported. These results contribute to significantly expanding genetic data available for African Saturniidae and allow for the development of new mitochondrial markers in future studies.
Keywords: Attacini; Bunaeini; Edible insects; Eochroini; Micragonini; Mitogenome; Phylogenetics; Saturniidae; Southern Africa.
©2022 Nethavhani et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures







References
-
- Barber J, Leavell BC, Keener AL, Breinholt JW, Chadwell BA, McClure CJW, Hill GM, Kawahara A. Moth tails divert bat attack: evolution of acoustic deflection. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2812–2816. doi: 10.1073/pnas.1421926112. - DOI - PMC - PubMed
-
- Bouvier E. Étude des Saturnioides normaux: famille des Saturniidés. Mémoires du Muséum National d’Histoire Naturelle (Nouvelle Série) 1936;3:1–354.
Further Reading
-
- Hong MY, Lee EM, Jo YH, Park HC, Kim SR, Hwang JS, Jin BR, Kang PD, Kim KG, Han YS, Kim I. Complete nucleotide sequence and organization of the mitogenome of the silk moth Caligula boisduvalii (Lepidoptera: Saturniidae) and comparison with other lepidopteran insects. Gene. 2008;413:49–57. doi: 10.1016/j.gene.2008.01.019. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

