IBD Subtype-Regulators IFNG and GBP5 Identified by Causal Inference Drive More Intense Innate Immunity and Inflammatory Responses in CD Than Those in UC
- PMID: 35462887
- PMCID: PMC9020454
- DOI: 10.3389/fphar.2022.869200
IBD Subtype-Regulators IFNG and GBP5 Identified by Causal Inference Drive More Intense Innate Immunity and Inflammatory Responses in CD Than Those in UC
Abstract
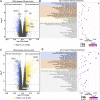
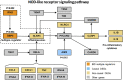
Background: The pathological differences between Crohn's disease (CD) and ulcerative colitis (UC) are substantial and unexplained yet. Here, we aimed to identify potential regulators that drive different pathogenesis of CD and UC by causal inference analysis of transcriptome data. Methods: Kruskal-Wallis and Dunnett's tests were performed to identify differentially expressed genes (DEGs) among CD patients, UC patients, and controls. Subsequently, differentially expressed pathways (DEPs) between CD and UC were identified and used to construct the interaction network of DEPs. Causal inference was performed to identify IBD subtype-regulators. The expression of the subtype-regulators and their downstream genes was validated by qRT-PCR with an independent cohort. Results: Compared with the control group, we identified 1,352 and 2,081 DEGs in CD and UC groups, respectively. Multiple DEPs between CD and UC were closely related to inflammation-related pathways, such as NOD-like receptor signaling, IL-17 signaling, and chemokine signaling pathways. Based on the priori interaction network of DEPs, causal inference analysis identified IFNG and GBP5 as IBD subtype-regulators. The results with the discovery cohort showed that the expression level of IFNG, GBP5, and NLRP3 was significantly higher in the CD group than that in the UC group. The regulation relationships among IFNG, GBP5, and NLRP3 were confirmed with transcriptome data from an independent cohort and validated by qRT-PCR. Conclusion: Our study suggests that IFNG and GBP5 were IBD subtype-regulators that trigger more intense innate immunity and inflammatory responses in CD than those in UC. Our findings reveal pathomechanical differences between CD and UC that may contribute to personalized treatment for CD and UC.
Keywords: Crohn’s disease; IBD subtype-regulator; causal inference; inflammatory bowel disease; ulcerative colitis.
Copyright © 2022 Gao, Li, Wu, Jiao, Yang, Zhao, Xu, Chen, Lin, Cheng, Zhu, Lan and Zhu.
Conflict of interest statement
Author RxZ serves as an advisory board member for Research Institute, GloriousMed Clinical Laboratory, GloriousMed Technology Co., Ltd., China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

