Extended-Spectrum ß-Lactamase-Producing Escherichia coli Among Humans, Beef Cattle, and Abattoir Environments in Nigeria
- PMID: 35463650
- PMCID: PMC9021871
- DOI: 10.3389/fcimb.2022.869314
Extended-Spectrum ß-Lactamase-Producing Escherichia coli Among Humans, Beef Cattle, and Abattoir Environments in Nigeria
Abstract
Introduction: Beef cattle, one of the food-producing animals, are linked to humans through a shared environment and the food chain as a major source of animal protein. Antimicrobial drugs are readily accessible for use in food animal production in Nigeria. Beef cattle and abattoir environments harbor pathogenic bacteria such as Escherichia coli (E. coli) which have developed resistance to antimicrobial agents used for prophylaxis or treatment. This study investigated the zoonotic transmission of extended-spectrum beta-lactamase-producing E. coli (ESBL-EC) among humans, beef cattle, and abattoir environments in Abuja and Lagos, Nigeria.
Materials and methods: We conducted a cross-sectional study among abattoir workers, beef cattle, and abattoir environments in Abuja and Lagos. Stool, cecal, and environmental samples were collected from apparently healthy workers, slaughtered cattle, and abattoir environments from May to December 2020. Data were collected electronically using open data kit app installed on a mobile phone. Antimicrobial susceptibility patterns were determined using the Kirby-Bauer disk diffusion method against a panel of 16 antimicrobial agents. Phenotypic and genotypic characterizations of the isolates were conducted. Data were analyzed with descriptive statistics.
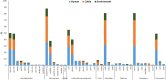
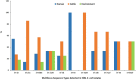
Results: From 21.7% (n = 97) of 448 samples, ESBL-EC were isolated and further characterized. Prevalence of ESBL-EC was highest in cattle (45.4%; n = 44), abattoir workers (41.2%; n = 40), and abattoir environment (13.4%; n = 13). Whole-genome sequencing of ESBL-EC showed dissemination of blaCTX-M-15 (90.7%; n = 88); blaCTX-M-14 (5.2%; n = 5); and blaCTX-M-55 (2.1%; n = 2) genes. The blaCTX-M-15 coexisted with blaCTX-M-14 and blaTEM-1 genes in 2.1% (n = 2) and 39.2% (n = 38) of the isolates, respectively. The presence of blaCTX-M-14 and blaCTX-M-15 genes was significantly associated with isolates originating from abattoir workers when compared with beef cattle isolates (p = 0.05; p < 0.01). The most prevalent sequence types (ST) were ST10 (n = 11), ST215 (n = 7), ST4684 (n = 7), and ST2178 (n = 6). ESBL-EC strain (ST205/B1) harbored mcr-1.1 and blaCTX-M15 and was isolated from a worker at Lagos abattoir. In 91 ESBL-EC isolates, 219 mobile genetic elements (MGEs) harbored resistance genes out of which β-lactam genes were carried on 64 different MGEs. Isolates showed equal distribution of insertion sequences and miniature inverted repeats although only a few composite transposons were detected (humans n = 12; cattle n = 9; environment n = 4). Two isolates of human and cattle origin (ST46/A) harboring ESBL genes and carried by MGEs were clonally related.
Conclusions: This is the first report of blaCTX-M-55 gene in humans and cattle in Nigeria. This study demonstrates the horizontal transfer of ESBL genes possibly by MGEs and buttresses the importance of genomic surveillance. Healthcare workers should be sensitized that people working closely with cattle or in abattoir environments are a high-risk group for fecal carriage of ESBL-EC when compared with the general population.
Keywords: Escherichia coli; Nigeria; abattoir; antimicrobial; cattle; environment; resistance.
Copyright © 2022 Aworh, Ekeng, Nilsson, Egyir, Owusu-Nyantakyi and Hendriksen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Agyare C., Etsiapa Boamah V., Ngofi Zumbi C., Boateng Osei F. (2019). “Antibiotic Use in Poultry Production and Its Effects on Bacterial Resistance,” in Antimicrobial Resistance - A Global Threat (InTech Open Science; ) doi: 10.5772/intechopen.79371 - DOI
-
- Bettencourt E. M. V., Tilman M., Narciso V., da Silva Carvalho M. L., de Sousa Henriques P. D. (2015). The Livestock Roles in the Wellbeing of Rural Communities of Timor-Leste. Rev. Econ e Sociol Rural 53, S063–S080. doi: 10.1590/1234-56781806-94790053s01005 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

