Integrated Bioinformatics-Based Analysis of Hub Genes and the Mechanism of Immune Infiltration Associated With Acute Myocardial Infarction
- PMID: 35463752
- PMCID: PMC9019083
- DOI: 10.3389/fcvm.2022.831605
Integrated Bioinformatics-Based Analysis of Hub Genes and the Mechanism of Immune Infiltration Associated With Acute Myocardial Infarction
Abstract
Background: Acute myocardial infarction (AMI) is a fatal disease that causes high morbidity and mortality. It has been reported that AMI is associated with immune cell infiltration. Now, we aimed to identify the potential diagnostic biomarkers of AMI and uncover the immune cell infiltration profile of AMI.
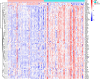
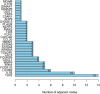
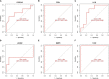
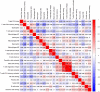
Methods: From the Gene Expression Omnibus (GEO) data set, three data sets (GSE48060, GSE60993, and GSE66360) were downloaded. Differentially expressed genes (DEGs) from AMI and healthy control samples were screened. Furthermore, DEGs were performed via gene ontology (GO) functional and kyoto encyclopedia of genes and genome (KEGG) pathway analyses. The Gene set enrichment analysis (GSEA) was used to analyze GO terms and KEGG pathways. Utilizing the Search Tool for Retrieval of Interacting Genes/Proteins (STRING) database, a protein-protein interaction (PPI) network was constructed, and the hub genes were identified. Then, the receiver operating characteristic (ROC) curves were constructed to analyze the diagnostic value of hub genes. And, the diagnostic value of hub genes was further validated in an independent data set GSE61144. Finally, CIBERSORT was used to represent the compositional patterns of the 22 types of immune cell fractions in AMI.
Results: A total of 71 DEGs were identified. These DEGs were mainly enriched in immune response and immune-related pathways. Toll-like receptor 2 (TLR2), interleukin-1B (IL1B), leukocyte immunoglobulin-like receptor subfamily B2 (LILRB2), Fc fragment of IgE receptor Ig (FCER1G), formyl peptide receptor 1 (FPR1), and matrix metalloproteinase 9 (MMP9) were identified as diagnostic markers with the value of p < 0.05. Also, the immune cell infiltration analysis indicated that TLR2, IL1B, LILRB2, FCER1G, FPR1, and MMP9 were correlated with neutrophils, monocytes, resting natural killer (NK) cells, gamma delta T cells, and CD4 memory resting T cells. The fractions of monocytes and neutrophils were significantly higher in AMI tissues than in control tissues.
Conclusion: TLR2, IL1B, LILRB2, FCER1G, FPR1, and MMP9 are involved in the process of AMI, which can be used as molecular biomarkers for the screening and diagnosis of AMI. In addition, the immune system plays a vital role in the occurrence and progression of AMI.
Keywords: CIBERSORT; acute myocardial infarction; bioinformatics; hub gene; immune cell infiltration.
Copyright © 2022 Wu, Jiang, Hua, Xiong, Chen, Li, Peng and Xiong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

