Prediction of Site Directed miRNAs as Key Players of Transcriptional Regulators Against Influenza C Virus Infection Through Computational Approaches
- PMID: 35463952
- PMCID: PMC9023806
- DOI: 10.3389/fmolb.2022.866072
Prediction of Site Directed miRNAs as Key Players of Transcriptional Regulators Against Influenza C Virus Infection Through Computational Approaches
Abstract
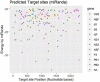
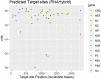
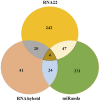
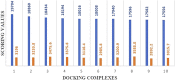
MicroRNAs (miRNAs) are small non-coding RNAs that play critical roles in gene expression, cell differentiation, and immunity against viral infections. In this study, we have used the computational tools, RNA22, RNAhybrid, and miRanda, to predict the microRNA-mRNA binding sites to find the putative microRNAs playing role in the host response to influenza C virus infection. This computational research screened the following four miRNAs: hsa-mir-3155a, hsa-mir-6796-5p, hsa-mir-3194-3p and hsa-mir-4673, which were further investigated for binding site prediction to the influenza C genome. Moreover, multiple sites in protein-coding region (HEF, CM2, M1-M2, NP, NS1- NS2, NSF, P3, PB1 and PB2) were predicted by RNA22, RNAhybrid and miRanda. Furthermore, 3D structures of all miRNAs and HEF were predicted and checked for their binding potential through molecular docking analysis. The comparative results showed that among all proteins, HEF is higher in prevalence throughout the analysis as a potential (human-derived) microRNAs target. The target-site conservation results showed that core nucleotide sequence in three different strains is responsible for potential miRNA binding to different viral strains. Further steps to use these microRNAs may lead to new therapeutic insights on fighting influenza virus infection.
Keywords: HEF; RNAComposer; RStudio; influenza C virus; miRNAs; mirbase; target site prediction.
Copyright © 2022 Hassan, Iqbal, Naqvi, Alashwal, Moustafa and Kloczkowski.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Ali Q., Jamal A., Ullah S., Waqar A. B. (2020). Male Predominant Association with Apolipoprotein B mRNA-Editing Enzyme, Catalytic Polypeptide-like 3G Variants (Rs6001417, Rs35228531, Rs8177832) Predict protection against HIV-1 Infection. Advancements Life Sci. 7 (2), 91–97.
-
- Asif M., Nawaz S., Bhutta Z. A., Kulyar M. F.-e.-A., Rashid M., Shabir S., et al. (2020). Viral Outbreaks: A Real Threat to the World. Advancements Life Sci. 8 (1), 08–19.
-
- Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008). The microRNA.Org Resource: Targets and Expression. Nucleic Acids Res. 36 (Suppl. l_1), D149–D153. 10.1093/nar/gkm995 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

