TNF Signaling Acts Downstream of MiR-322/-503 in Regulating DM1 Myogenesis
- PMID: 35464065
- PMCID: PMC9021394
- DOI: 10.3389/fendo.2022.843202
TNF Signaling Acts Downstream of MiR-322/-503 in Regulating DM1 Myogenesis
Abstract
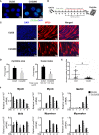
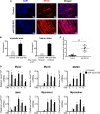
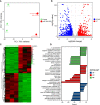
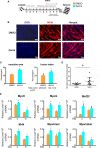
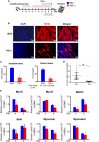
Myotonic dystrophy type 1 (DM1) is caused by the expanded CUG repeats and usually displays defective myogenesis. Although we previously reported that ectopic miR-322/-503 expression improved myogenesis in DM1 by targeting the toxic RNA, the underlying pathways regulating myogenesis that were aberrantly altered in DM1 and rescued by miR-322/-503 were still unknown. Here, we constructed DM1 and miR-322/-503 overexpressing DM1 myoblast models, which were subjected to in vitro myoblast differentiation along with their corresponding controls. Agreeing with previous findings, DM1 myoblast showed remarkable myogenesis defects, while miR-322/-503 overexpression successfully rescued the defects. By RNA sequencing, we noticed that Tumor necrosis factor (TNF) signaling was the only pathway that was significantly and oppositely altered in these two experimental sets, with it upregulated in DM1 and inhibited by miR-322/-503 overexpression. Consistently, hyperactivity of TNF signaling was detected in two DM1 mouse models. Blocking TNF signaling significantly rescued the myogenesis defects in DM1. On the contrary, TNF-α treatment abolished the rescue effect of miR-322/-503 on DM1 myogenesis. Taking together, these results implied that TNF signaling mediated the myogenesis defects in DM1 and might act downstream of miR-322/-503 in regulating the myogenesis in DM1. Moreover, the inhibition of TNF signaling benefiting myogenesis in DM1 provided us with a novel therapeutic strategy for DM1.
Keywords: DM1; TNF signaling; miR-322/-503; myoblast; myogenesis.
Copyright © 2022 Li, Xu, Liu, Wang, Zhao, Zhu and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

