Comprehensive Oncogenic Features of Coronavirus Receptors in Glioblastoma Multiforme
- PMID: 35464443
- PMCID: PMC9020264
- DOI: 10.3389/fimmu.2022.840785
Comprehensive Oncogenic Features of Coronavirus Receptors in Glioblastoma Multiforme
Abstract
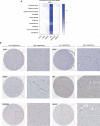
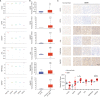
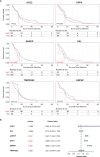
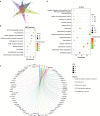
The COVID-19 pandemic caused by SARS-CoV-2 infection has placed health systems under excessive pressure and especially elderly people with cancer. Glioblastoma multiforme (GBM) is a malignant brain tumor with an increasing incidence in elderly individuals, and thereby GBM patients are a vulnerable population during the COVID-19 outbreak. Accumulating studies have implied that SARS-CoV-2 might invade the brain directly via coronavirus receptors. However, little is known about SARS-CoV-2 infection in the clinical development of GBM. Here, we explored the oncogenic roles of six coronavirus receptors (ACE2, DPP4, ANPEP, AXL, TMPRSS2, and ENPEP) in GBM using bioinformatics and experimental approaches. We found that ANPEP and ENPEP were significantly increased at both the mRNA and protein levels in GBM compared with normal brain tissue. Kaplan-Meier survival curves and Cox regression analysis demonstrated that high expressions of ANPEP and ENPEP are associated with poor prognosis and survival. Moreover, all receptors are positively correlated with the immune infiltration levels of monocyte. Furthermore, we identified 245 genes between COVID-19 and coronavirus receptors-correlated genes in GBM and performed a thorough analysis of their protein-protein interaction network, functional signaling pathway and molecular process. Our work explores for the first time the association of coronavirus receptors with GBM and suggests ANPEP and ENPEP as potential therapeutic targets of GBM irrespective of COVID-19.
Keywords: ACE2; ANPEP; ENPEP; GBM; SARS-CoV-2.
Copyright © 2022 Chen, Zhao, Li, Sun, Ma, Peng, Shi, Li and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses.Biochem Biophys Res Commun. 2020 May 21;526(1):135-140. doi: 10.1016/j.bbrc.2020.03.044. Epub 2020 Mar 19. Biochem Biophys Res Commun. 2020. PMID: 32199615 Free PMC article.
-
Evidence for residual SARS-CoV-2 in glioblastoma tissue of a convalescent patient.Neuroreport. 2021 Jun 9;32(9):771-775. doi: 10.1097/WNR.0000000000001654. Neuroreport. 2021. PMID: 33994523
-
Bioinformatic characterization of ENPEP, the gene encoding a potential cofactor for SARS-CoV-2 infection.PLoS One. 2024 Dec 11;19(12):e0307731. doi: 10.1371/journal.pone.0307731. eCollection 2024. PLoS One. 2024. PMID: 39661628 Free PMC article.
-
Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19).J Pathol. 2020 Jul;251(3):228-248. doi: 10.1002/path.5471. Epub 2020 Jun 10. J Pathol. 2020. PMID: 32418199 Free PMC article. Review.
-
Polymorphisms and mutations of ACE2 and TMPRSS2 genes are associated with COVID-19: a systematic review.Eur J Med Res. 2022 Feb 22;27(1):26. doi: 10.1186/s40001-022-00647-6. Eur J Med Res. 2022. PMID: 35193695 Free PMC article.
Cited by
-
Evaluating glioblastoma tumour sphere growth and migration in interaction with astrocytes using 3D collagen-hyaluronic acid hydrogels.J Mater Chem B. 2023 Jun 21;11(24):5442-5459. doi: 10.1039/d3tb00066d. J Mater Chem B. 2023. PMID: 37159233 Free PMC article.
-
COVID-19 hospitalization increases the risk of developing glioblastoma: a bidirectional Mendelian-randomization study.Front Oncol. 2023 Aug 21;13:1185466. doi: 10.3389/fonc.2023.1185466. eCollection 2023. Front Oncol. 2023. PMID: 37671050 Free PMC article.
-
Excavation of gene markers associated with pancreatic ductal adenocarcinoma based on interrelationships of gene expression.IET Syst Biol. 2024 Dec;18(6):261-270. doi: 10.1049/syb2.12090. Epub 2024 Mar 26. IET Syst Biol. 2024. PMID: 38530028 Free PMC article.
-
Signaling Pathways of AXL Receptor Tyrosine Kinase Contribute to the Pathogenetic Mechanisms of Glioblastoma.Cells. 2024 Feb 19;13(4):361. doi: 10.3390/cells13040361. Cells. 2024. PMID: 38391974 Free PMC article. Review.
-
Angiotensin‑converting enzyme 2 expression in human tumors: Implications for prognosis and therapy (Review).Oncol Rep. 2025 Sep;54(3):101. doi: 10.3892/or.2025.8934. Epub 2025 Jun 27. Oncol Rep. 2025. PMID: 40576111 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

