Identification and Validation of Chromobox Family Members as Potential Prognostic Biomarkers and Therapeutic Targets for Human Esophageal Cancer
- PMID: 35464847
- PMCID: PMC9019303
- DOI: 10.3389/fgene.2022.851390
Identification and Validation of Chromobox Family Members as Potential Prognostic Biomarkers and Therapeutic Targets for Human Esophageal Cancer
Abstract
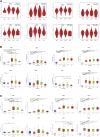
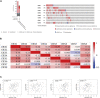
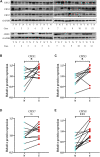
Background: Chromobox family proteins (CBXs) are vital components of epigenetic regulation complexes and transcriptionally inhibit target genes by modifying the chromatin. Accumulating evidence indicates that CBXs are involved in the initiation and progression of multiple malignancies. However, the expression, function, and clinical relevance such as the prognostic and diagnostic values of different CBXs in esophageal carcinoma (ESCA) are still unclear. Methods: We applied Oncomine, TCGA, GEO, GEPIA, UALCAN, Kaplan-Meier plotter, cBioPortal, Metascape, and TIMER to investigate the roles of CBX family members in ESCA. Additionally, quantitative real-time PCR (RT-PCR), western blot, and immunofluorescence were used to verify the expression of CBX family members in ESCA clinical samples. Results: Compared with normal tissues, the mRNA expression levels of CBX1/3/8 were significantly increased in ESCA, whereas CBX7 mRNA expression was reduced in both the TCGA cohort and GEO cohort. In the TCGA cohort, ROC curves suggested that CBX1/2/3/4/8 had great diagnostic value in ESCA, and the AUCs were above 0.9. Furthermore, upregulation of CBX1/3/8 and downregulation of CBX7 were closely related to the clinicopathological parameters in ESCA patients, such as tumor grades, tumor nodal metastasis status, and TP53 mutation status. The survival analysis indicated that higher CBX1/3/8 mRNA expressions and lower CBX7 expression suggested an unfavorable prognosis in ESCA. High genetic change rate (52%) of CBXs was found in ESCA patients. Functions and pathways of mutations in CBXs and their 50 frequently altered neighbor genes in ESCA patients were investigated; the results showed that DNA repair and DNA replication were correlated to CBX alterations. Moreover, we found a significant correlation between the expression level of CBX family members and the infiltration of immune cells in ESCA. Finally, we verified the expression of CBX family members in clinical samples and found the results were consistent with the databases. Conclusion: Our study implied that CBX1/3/7/8 are potential targets of precision therapy for ESCA patients and new biomarkers for the prognosis.
Keywords: bioinformatics analysis; biomarker; chromobox (CBX) protein; esophageal cancer; immunofluorescence; prognosis.
Copyright © 2022 Fang, Wang, Chen, Zhuang, Huang, Chen, Huang, Zheng and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., et al. (2012). The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2 (5), 401–404. 10.1158/2159-8290.CD-12-0095 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

