A Comprehensive Analysis of Citrus Tristeza Variants of Bhutan and Across the World
- PMID: 35464978
- PMCID: PMC9024366
- DOI: 10.3389/fmicb.2022.797463
A Comprehensive Analysis of Citrus Tristeza Variants of Bhutan and Across the World
Abstract
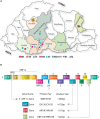
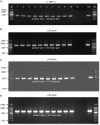
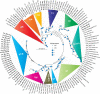
Mandarin orange is economically one of the most important fruit crops in Bhutan. However, in recent years, orange productivity has dropped due to severe infection of citrus tristeza virus (CTV) associated with the gradual decline of citrus orchards. Although the disease incidence has been reported, very limited information is available on genetic variability among the Bhutanese CTV variants. This study used reverse transcription PCR (RT-PCR) to detect CTV in collected field samples and recorded disease incidence up to 71.11% in Bhutan's prominent citrus-growing regions. To elucidate the extent of genetic variabilities among the Bhutanese CTV variants, we targeted four independent genomic regions (5'ORF1a, p25, p23, and p18) and analyzed a total of 64 collected isolates. These genomic regions were amplified and sequenced for further comparative bioinformatics analysis. Comprehensive phylogenetic reconstructions of the GenBank deposited sequences, including the corresponding genomic locations from 53 whole-genome sequences, revealed unexpected and rich diversity among Bhutanese CTV variants. A resistant-breaking (RB) variant was also identified for the first time from the Asian subcontinent. Our analyses unambiguously identified five (T36, T3, T68, VT, and HA16-5) major, well-recognized CTV strains. Bhutanese CTV variants form two additional newly identified distinct clades with higher confidence, B1 and B2, named after Bhutan. The origin of each of these nine clades can be traced back to their root in the north-eastern region of India and Bhutan. Together, our study established a definitive framework for categorizing global CTV variants into their distinctive clades and provided novel insights into multiple genomic region-based genetic diversity assessments, including their pathogenicity status.
Keywords: RT-PCR; citrus tristeza virus; genomic diversity; genomic regions; sequencing and phylogenetic analysis.
Copyright © 2022 Ghosh, Kokane, Kokane, Mukherjee, Tenzin, Surwase, Deshmukh, Gubyad and Biswas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Detection and Molecular Characterization of 'Candidatus Liberibacter asiaticus' and Citrus Tristeza Virus Associated with Citrus Decline in Bhutan.Phytopathology. 2021 May;111(5):870-881. doi: 10.1094/PHYTO-07-20-0266-R. Epub 2021 Apr 14. Phytopathology. 2021. PMID: 33090079
-
Distribution, genetic diversity and recombination analysis of Citrus tristeza virus of India.Virus Genes. 2012 Aug;45(1):139-48. doi: 10.1007/s11262-012-0748-3. Epub 2012 May 5. Virus Genes. 2012. PMID: 22562224
-
Population structure and diversity of citrus tristeza virus (CTV) isolates in Hunan province, China.Arch Virol. 2017 Feb;162(2):409-423. doi: 10.1007/s00705-016-3089-z. Epub 2016 Oct 22. Arch Virol. 2017. PMID: 27771790
-
Serological and Molecular Detection of Citrus Tristeza Virus: A Review.Microorganisms. 2024 Jul 27;12(8):1539. doi: 10.3390/microorganisms12081539. Microorganisms. 2024. PMID: 39203383 Free PMC article. Review.
-
Walking Together: Cross-Protection, Genome Conservation, and the Replication Machinery of Citrus tristeza virus.Viruses. 2020 Nov 26;12(12):1353. doi: 10.3390/v12121353. Viruses. 2020. PMID: 33256049 Free PMC article. Review.
Cited by
-
Huanglongbing Pandemic: Current Challenges and Emerging Management Strategies.Plants (Basel). 2022 Dec 29;12(1):160. doi: 10.3390/plants12010160. Plants (Basel). 2022. PMID: 36616289 Free PMC article. Review.
-
Development of diagnostic tools and discovery of two novel Indian citrus ringspot virus species: insights into global mandarivirus phylogeography.Front Microbiol. 2025 Feb 14;16:1513291. doi: 10.3389/fmicb.2025.1513291. eCollection 2025. Front Microbiol. 2025. PMID: 40028458 Free PMC article.
-
Variation of endosymbiont and citrus tristeza virus (CTV) titers in the Huanglongbing insect vector, Diaphorina citri, on CTV-infected plants.Front Microbiol. 2023 Sep 22;14:1236731. doi: 10.3389/fmicb.2023.1236731. eCollection 2023. Front Microbiol. 2023. PMID: 37808301 Free PMC article.
References
-
- Ahlawat Y. S. (2012). Virus Diseases of Citrus & Management. New Delhi: Studium Press (India).
LinkOut - more resources
Full Text Sources

