Analysis of Interaction Network Between Host Protein and M Protein of Swine Acute Diarrhea Syndrome Coronavirus
- PMID: 35464981
- PMCID: PMC9024367
- DOI: 10.3389/fmicb.2022.858460
Analysis of Interaction Network Between Host Protein and M Protein of Swine Acute Diarrhea Syndrome Coronavirus
Abstract
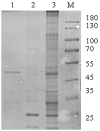
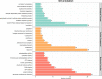
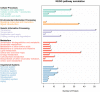
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is an enterovirus that can cause acute diarrhea and death in piglets and cause serious economic losses to the pig industry. SADS-CoV membrane (M) protein mainly plays a key role in biological processes, such as virus assembly, budding, and host innate immune regulation. Understanding the interaction between M protein and host proteins is very important to define the molecular mechanism of cells at the protein level and to understand specific cellular physiological pathways. In this study, 289 host proteins interacting with M protein were identified by glutathione-S-transferase (GST) pull-down combined with liquid chromatography-mass spectrometry (LC-MS/MS), and the protein-protein interaction (PPI) network was established by Gene Ontology (GO) terms and Kyoto Encyclopedia of Gene and Genomes (KEGG) pathways analysis. Results showed that SADS-CoV M protein was mainly associated with the host metabolism, signal transduction, and innate immunity. The Co-Immunoprecipitation (CO-IP) validation results of six randomly selected proteins, namely, Rab11b, voltage-dependent anion-selective channel 1 (VDAC1), Ribosomal Protein L18 (RPL18), RALY, Ras Homolog Family Member A (RHOA), and Annexin A2 (ANXA2), were consistent with LC-MS results. In addition, overexpression of RPL18 and PHOA significantly promoted SADS-CoV replication, while overexpression of RALY antagonized viral replication. This work will help to clarify the function of SADS-CoV M protein in the life cycle of SADS-CoV.
Keywords: RALY; RHOA; RPL18; membrane (M) protein; protein interaction network; swine acute diarrhea syndrome coronavirus (SADS-CoV).
Copyright © 2022 Xu, Cao, Ji, Zhou, Yan, Sun and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

