Mechanism of reaction of RNA-dependent RNA polymerase from SARS-CoV-2
- PMID: 35465139
- PMCID: PMC9016896
- DOI: 10.1016/j.checat.2022.03.019
Mechanism of reaction of RNA-dependent RNA polymerase from SARS-CoV-2
Abstract
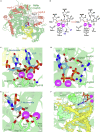
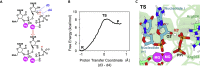
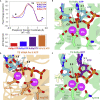
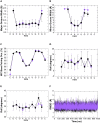
We combine molecular dynamics, statistical mechanics, and hybrid quantum mechanics/molecular mechanics simulations to describe mechanistically the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp). Our study analyzes the binding mode of both natural triphosphate substrates as well as remdesivir triphosphate (the active form of drug), which is bound preferentially over ATP by RdRp while being poorly recognized by human RNA polymerase II (RNA Pol II). A comparison of incorporation rates between natural and antiviral nucleotides shows that remdesivir is incorporated more slowly into the nascent RNA compared with ATP, leading to an RNA duplex that is structurally very similar to an unmodified one, arguing against the hypothesis that remdesivir is a competitive inhibitor of ATP. We characterize the entire mechanism of reaction, finding that viral RdRp is highly processive and displays a higher catalytic rate of incorporation than human RNA Pol II. Overall, our study provides the first detailed explanation of the replication mechanism of RdRp.
Keywords: QM/MM; RNA-dependent RNA polymerase; SARS-CoV-2; antivirals; biocatalysis; free energy calculations; reaction mechanism; remdesivir; viral replication.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- WHO . WHO; 2021. Novel Coronavirus (COVID-19) Situation.
-
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Genomic epidemiology of novel coronavirus - global subsampling. Nextstrain real-time Track. Pathog. Evol. 2020;26:2854.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
