P2X3-selective mechanism of Gefapixant, a drug candidate for the treatment of refractory chronic cough
- PMID: 35465163
- PMCID: PMC9014320
- DOI: 10.1016/j.csbj.2022.03.030
P2X3-selective mechanism of Gefapixant, a drug candidate for the treatment of refractory chronic cough
Abstract
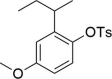
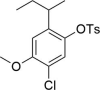
Gefapixant/AF-219, a selective inhibitor of the P2X3 receptor, is the first new drug other than dextromethorphan to be approved for the treatment of refractory chronic cough (RCC) in nearly 60 years. To date, seven P2X subtypes (P2X1-7) activated by extracellular ATP have been cloned, and subtype selectivity of P2X inhibitors is a prerequisite for reducing side effects. We previously identified the site and mechanism of action of Gefapixant/AF-219 on the P2X3 receptor, which occupies a pocket consisting of the left flipper (LF) and lower body (LB) domains. However, the mechanism by which AF-219 selectively acts on the P2X3 receptor is unknown. Here, we combined mutagenesis, chimera construction, molecular simulations, covalent occupation and chemical synthesis, and find that the negative allosteric site of AF-219 at P2X3 is also present in other P2X subtypes, at least for P2X1, P2X2 and P2X4. By constructing each chimera of AF-219 sensitive P2X3 and insensitive P2X2 subtypes, the insensitive P2X2 subtype was made to acquire the inhibitory properties of AF-219 and AF-353, an analog of AF-219 with higher affinity. Our results suggest that the selectivity of AF-219/AF-353 for P2X3 over the other P2X subtypes is determined by a combination of the accessibility of P2X3 binding site and the internal shape of this pocket, a finding that could provide new perspectives for drug design against P2X3-mediated diseases such as RCC, idiopathic pulmonary fibrosis, hypertension and overactive bladder disorder.
Keywords: Binding sites; DF, dorsal fin; Gefapixant/AF-219; IC50, the concentration yielding half of the maximal inhibition; LB, lower body; LF, left flipper; MD, molecular dynamics; NPM, N-phenylmaleimide; P2X3 receptors; RCC, refractory chronic cough; Refractory chronic cough; Subtype selectivity.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures













References
-
- Burnstock G. The therapeutic potential of purinergic signalling. Biochem Pharmacol. 2018;151:157–165. - PubMed
-
- Muccino D., Green S. Update on the clinical development of gefapixant, a P2X3 receptor antagonist for the treatment of refractory chronic cough. Pulm Pharmacol Ther. 2019;56:75–78. - PubMed
-
- Morice A.H., Kitt M.M., Ford A.P., Tershakovec A.M., Wu W.C., Brindle K., et al. The effect of gefapixant, a P2X3 antagonist, on cough reflex sensitivity: a randomised placebo-controlled study. Eur Respir J. 2019;54 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
