Chemical-induced gene expression ranking and its application to pancreatic cancer drug repurposing
- PMID: 35465231
- PMCID: PMC9023899
- DOI: 10.1016/j.patter.2022.100441
Chemical-induced gene expression ranking and its application to pancreatic cancer drug repurposing
Abstract
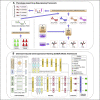
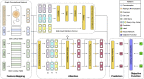
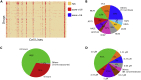
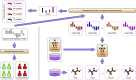
Chemical-induced gene expression profiles provide critical information of chemicals in a biological system, thus offering new opportunities for drug discovery. Despite their success, large-scale analysis leveraging gene expressions is limited by time and cost. Although several methods for predicting gene expressions were proposed, they only focused on imputation and classification settings, which have limited applications to real-world scenarios of drug discovery. Therefore, a chemical-induced gene expression ranking (CIGER) framework is proposed to target a more realistic but more challenging setting in which overall rankings in gene expression profiles induced by de novo chemicals are predicted. The experimental results show that CIGER significantly outperforms existing methods in both ranking and classification metrics. Furthermore, a drug screening pipeline based on CIGER is proposed to identify potential treatments of drug-resistant pancreatic cancer. Our predictions have been validated by experiments, thereby showing the effectiveness of CIGER for phenotypic compound screening of precision medicine.
Keywords: attention; cancer therapy; drug repurposing; gene expression; graph neural network; learning-to-rank; machine learning; pancreatic cancer; phenotype screening; precision medicine.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Comment in
-
AI in drug discovery: Applications, opportunities, and challenges.Patterns (N Y). 2022 Jun 10;3(6):100529. doi: 10.1016/j.patter.2022.100529. eCollection 2022 Jun 10. Patterns (N Y). 2022. PMID: 35755871 Free PMC article. No abstract available.
References
-
- Terstappen G.C., Schlüpen C., Raggiaschi R., Gaviraghi G. Target deconvolution strategies in drug discovery. Nat. Rev. Drug Discov. 2007;6:891–903. - PubMed
-
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.-P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

