Molecular analysis of TCGA breast cancer histologic types
- PMID: 35465400
- PMCID: PMC9028992
- DOI: 10.1016/j.xgen.2021.100067
Molecular analysis of TCGA breast cancer histologic types
Abstract
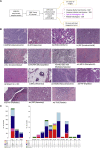
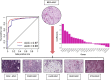
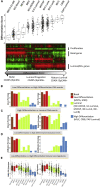
Breast cancer is classified into multiple distinct histologic types, and many of the rarer types have limited characterization. Here, we extend The Cancer Genome Atlas Breast Cancer (TCGA-BRCA) dataset with additional histologic type annotations, in a total of 1063 breast cancers. We analyze this extended dataset to define transcriptomic and genomic profiles of six rare special histologic types: cribriform, micropapillary, mucinous, papillary, metaplastic, and invasive carcinoma with medullary pattern. We show the broader applicability of our constructed special histologic type gene signatures in the TCGA Pan-Cancer Atlas dataset with a predictive model that detects mucinous histologic type across cancers of other organ systems. Using a normal mammary cell differentiation score analysis, we order histologic types into a continuum from stem cell-like to luminal progenitor-like to mature luminal-like. Finally, we classify TCGA-BRCA into 12 consensus groups based on integrated genomic and histological features. We present a rich openly accessible resource of histologic and genomic characterization of TCGA-BRCA to enable studies of the range of breast cancers.
Conflict of interest statement
C.M.P. is an equity stockholder and consultant of BioClassifier LLC; C.M.P. is also listed as an inventor on patent applications for the Breast PAM50 Subtyping assay.
Figures



References
-
- Tan P.H., Ellis I., Allison K., Brogi E., Fox S.B., Lakhani S., Lazar A.J., Morris E.A., Sahin A., Salgado R., et al. WHO Classification of Tumours Editorial Board The 2019 World Health Organization classification of tumours of the breast. Histopathology. 2020;77:181–185. doi: 10.1111/his.14091. - DOI - PubMed
-
- WHO . WHO Classification of Tumours. Fifth Edition. Volume 2. WHO; 2019. Breast Tumours.
-
- Weigelt B., Horlings H.M., Kreike B., Hayes M.M., Hauptmann M., Wessels L.F., de Jong D., Van de Vijver M.J., Van’t Veer L.J., Peterse J.L. Refinement of breast cancer classification by molecular characterization of histological special types. J. Pathol. 2008;216:141–150. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

