Long-Term Prediction of Blood Glucose Levels in Type 1 Diabetes Using a CNN-LSTM-Based Deep Neural Network
- PMID: 35466701
- PMCID: PMC10658677
- DOI: 10.1177/19322968221092785
Long-Term Prediction of Blood Glucose Levels in Type 1 Diabetes Using a CNN-LSTM-Based Deep Neural Network
Abstract
Background: In this work, we leverage state-of-the-art deep learning-based algorithms for blood glucose (BG) forecasting in people with type 1 diabetes.
Methods: We propose stacks of convolutional neural network and long short-term memory units to predict BG level for 30-, 60-, and 90-minute prediction horizon (PH), given historical glucose measurements, meal information, and insulin intakes. The evaluation was performed on two data sets, Replace-BG and DIAdvisor, representative of free-living conditions and in-hospital setting, respectively.
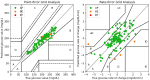
Results: For 90-minute PH, our model obtained mean absolute error of 17.30 ± 2.07 and 18.23 ± 2.97 mg/dL, root mean square error of 23.45 ± 3.18 and 25.12 ± 4.65 mg/dL, coefficient of determination of 84.13 ± 4.22% and 82.34 ± 4.54%, and in terms of the continuous glucose-error grid analysis 94.71 ± 3.89% and 91.71 ± 4.32% accurate predictions, 1.81 ± 1.06% and 2.51 ± 0.86% benign errors, and 3.47 ± 1.12% and 5.78 ± 1.72% erroneous predictions, for Replace-BG and DIAdvisor data sets, respectively.
Conclusion: Our investigation demonstrated that our method achieved superior glucose forecasting compared with existing approaches in the literature, and thanks to its generalizability showed potential for real-life applications.
Keywords: artificial deep neural networks; continuous glucose monitoring (CGM); convolutional neural network (CNN); decision support systems; glucose forecasting; long short-term memory (LSTM).
Conflict of interest statement
Declaration of Conflicting InterestsThe author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Dr Cescon serves on the advisory board for Diatech Diabetes, Inc. Mehrad Jaloli declares no conflict of interest relevant to this project.
Figures





References
-
- Kirchsteiger H, Jørgensen JB, Renard E, del Re L. Prediction Methods for Blood Glucose Concentration. Switzerland: Springer International Publishing. 2016.
-
- Sparacino G, Zanderigo F, Corazza S, Maran A, Facchinetti A, Cobelli C. Glucose concentration can be predicted ahead in time from continuous glucose monitoring sensor time-series. IEEE Trans Biomed Eng. 2007;54:931-937. - PubMed
-
- Gani A, Gribok AV, Lu Y, Ward WK, Vigersky RA, Reifman J. Universal glucose models for predicting subcutaneous glucose concentration in humans. IEEE Trans Inf Technol Biomed. 2009;14:157-165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

