Cutaneous Microbiome Profiles Following Chlorhexidine Treatment in a 72-Hour Daily Follow-Up Paired Design: a Pilot Study
- PMID: 35467392
- PMCID: PMC9248901
- DOI: 10.1128/spectrum.01753-21
Cutaneous Microbiome Profiles Following Chlorhexidine Treatment in a 72-Hour Daily Follow-Up Paired Design: a Pilot Study
Abstract
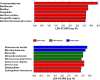
Venous catheter-related bloodstream infections represent a significant problem in the United States. Our objective was to determine daily changes in skin microbiome profiles up to 72h postchlorhexidine treatment. Left and right forearm skin swab samples were obtained from 10 healthy volunteers over 72h at 24h intervals. Dorsal surface of left arm was treated with chlorohexidine gluconate (CHG) at initial time point (T = 0), while the right arm remained untreated (control). Swab samples were obtained shortly before (T = 0) and after CHG treatment (T = 24-48-72h). Bacterial DNA extraction, 16S rRNA gene V1-V3 sequencing and taxonomic annotation were performed using ZymoBIOMICS pipeline. PERMANOVA, linear discriminant and bacterial interaction network analyses were performed. A total of 13 total phyla, 273 genera, and 950 total species were detected across all time points, CHG-treated or CHG-untreated. Most abundant species included Cutibacterium acnes, Staphylococcus epidermidis, and Rothia Mucilaginosa. Low biomass-related inconsistent taxa detection was observed. PERMANOVA suggested a marginal difference between CHG-treated and CHG-untreated microbiome profiles (Genera: P(perm) = 0.0531; Species: P(perm) = 0.0450). Bacterial interaction network guided PERMANOVA analyses detected a microbiome change over time, suggesting a consistent CHG treatment-specific change. LEfSe identified Finegoldia magna, Bacillus pumilus, Bacillus thermoamylovorans as the only distinctive species. These species were more abundant and/or present post-CHG treatment in the CHG-treated group. These findings suggest that the skin microbiome was not significantly different 24, 48, or 72h after CHG treatment. Previous culture-based studies have found similar results after 24h. Future studies will be needed to determine the mechanisms of bacterial regrowth after CHG treatment. IMPORTANCE Annually, over 80,000 central line infections occur in the United States. Understanding the pathogenesis of these infections is crucial. Chlorhexidine is the most commonly used skin preparation before line placement. We hypothesized that the use of chlorhexidine and dressings will alter the normal arm skin microbiome over a period of 72h. We used 16S-rRNA gene next generation sequencing (NGS) to determine the forearm skin microbiome of volunteers. The left arm was swabbed with chlorhexidine and the right arm served as control. The skin microbiome returned to normal after 24h. Our NGS results confirm findings of two previous culture-based studies. Relative abundance of Bacillus spp. in the chlorhexidine-treated samples was increased, consistent with one previous study. Based on the results of this pilot study, we will need to measure viable bacteria during a 24h time course following chlorhexidine treatment to understand the source of skin microbiome replenishment.
Keywords: 16S rRNA gene sequencing; bloodstream infections; chlorhexidine; skin microbiome.
Conflict of interest statement
The authors declare no conflict of interest. Cardinal Health partially funded the project.
Figures
References
-
- Mermel LA, Allon M, Bouza E, Craven DE, Flynn P, O'Grady NP, Raad II, Rijnders BJ, Sherertz RJ, Warren DK. 2009. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 Update by the Infectious Diseases Society of America. Clin Infect Dis 49:1–45. doi: 10.1086/599376. - DOI - PMC - PubMed
-
- Choudhury MA, Sidjabat HE, Zowawi HM, Marsh PhD N, Larsen E, Runnegar PhD N, Paterson DL, McMillan DJ, Rickard CM. 2019. Skin colonization at peripheral intravenous catheter insertion sites increases the risk of catheter colonization and infection. Am J Infect Control 47:1484–1488. doi: 10.1016/j.ajic.2019.06.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

