Construction and Analysis of the Complete Genome Sequence of Leprosy Agent Mycobacterium lepromatosis
- PMID: 35467405
- PMCID: PMC9248898
- DOI: 10.1128/spectrum.01692-21
Construction and Analysis of the Complete Genome Sequence of Leprosy Agent Mycobacterium lepromatosis
Abstract
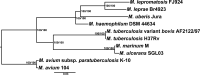
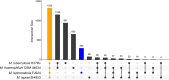
Leprosy is caused by Mycobacterium leprae and Mycobacterium lepromatosis. We report construction and analyses of the complete genome sequence of M. lepromatosis FJ924. The genome contained 3,271,694 nucleotides to encode 1,789 functional genes and 1,564 pseudogenes. It shared 1,420 genes and 885 pseudogenes (71.4%) with M. leprae but differed in 1,281 genes and pseudogenes (28.6%). In phylogeny, the leprosy bacilli started from a most recent common ancestor (MRCA) that diverged ~30 million years ago (Mya) from environmental organism Mycobacterium haemophilum. The MRCA then underwent reductive evolution with pseudogenization, gene loss, and chromosomal rearrangements. Analysis of the shared pseudogenes estimated the pseudogenization event ~14 Mya, shortly before species bifurcation. Afterwards, genomic changes occurred to lesser extent in each species. Like M. leprae, four major types of highly repetitive sequences were detected in M. lepromatosis, contributing to chromosomal rearrangements within and after MRCA. Variations in genes and copy numbers were noted, such as three copies of the gene encoding bifunctional diguanylate cyclase/phosphodiesterase in M. lepromatosis, but single copy in M. leprae; 6 genes encoding the TetR family transcriptional regulators in M. lepromatosis, but 11 such genes in M. leprae; presence of hemW gene in M. lepromatosis, but absence in M. leprae; and others. These variations likely aid unique pathogenesis, such as diffuse lepromatous leprosy associated with M. lepromatosis, while the shared genomic features should explain the common pathogenesis of dermatitis and neuritis in leprosy. Together, these findings and the genomic data of M. lepromatosis may facilitate future research and care for leprosy. IMPORTANCE Leprosy is a dreaded infection that still affects millions of people worldwide. Mycobacterium lepromatosis is a recently recognized cause in addition to the well-known Mycobacterium leprae. M. lepromatosis is likely specific for diffuse lepromatous leprosy, a severe form of the infection and endemic in Mexico. This study constructed and annotated the complete genome sequence of M. lepromatosis FJ924 and performed comparative genomic analyses with related mycobacteria. The results afford new and refined insights into the genome size, gene repertoire, pseudogenes, phylogenomic relationship, genome organization and plasticity, process and timing of reductive evolution, and genetic and proteomic basis for pathogenesis. The availability of the complete M. lepromatosis genome may prove to be useful for future research and care for the infection.
Keywords: Mycobacterium leprae; Mycobacterium lepromatosis; genomics; leprosy; reductive evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
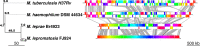
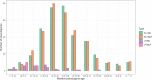


References
-
- Gelber RH. 2005. Leprosy (Hansen’s Disease). P 966–972. In Kasper DL, Braunwald E, Fauci AS, Hauser SL, Longo DL, Jameson JL (ed), Harrison’s principles of internal medicine, 16th ed McGraw-Hill, New York.
-
- Latapí F, Chevez Zamora A. 1948. The “spotted” leprosy of Lucio: an introduction to its clinical and histological study. Int J Lepr 16:421–430.
-
- Hansen GHA. 1874. Undersøgelser angående spedalskhedens årsager. Nor Mag Laegevidenskaben 9:1–88.
-
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011. doi:10.1038/35059006. - DOI - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

