Structure and function of MuvB complexes
- PMID: 35468940
- PMCID: PMC9201786
- DOI: 10.1038/s41388-022-02321-x
Structure and function of MuvB complexes
Abstract
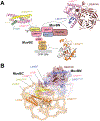
Proper progression through the cell-division cycle is critical to normal development and homeostasis and is necessarily misregulated in cancer. The key to cell-cycle regulation is the control of two waves of transcription that occur at the onset of DNA replication (S phase) and mitosis (M phase). MuvB complexes play a central role in the regulation of these genes. When cells are not actively dividing, the MuvB complex DREAM represses G1/S and G2/M genes. Remarkably, MuvB also forms activator complexes together with the oncogenic transcription factors B-MYB and FOXM1 that are required for the expression of the mitotic genes in G2/M. Despite this essential role in the control of cell division and the relationship to cancer, it has been unclear how MuvB complexes inhibit and stimulate gene expression. Here we review recent discoveries of MuvB structure and molecular interactions, including with nucleosomes and other chromatin-binding proteins, which have led to the first mechanistic models for the biochemical function of MuvB complexes.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests
The authors declare no competing interest.
Figures


References
-
- Morgan DO. The cell cycle : principles of control. Published by New Science Press in association with Oxford University Press; Distributed inside North America by Sinauer Associates, Publishers: London Sunderland, MA, 2007.
-
- Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res 2000; 60: 3689–3695. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. - PubMed
-
- Fischer M, Muller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev Biochem Mol Biol 2017; 52: 638–662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

