Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment
- PMID: 35469019
- PMCID: PMC9064804
- DOI: 10.1038/s41564-022-01091-2
Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment
Erratum in
-
Publisher Correction: Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment.Nat Microbiol. 2022 Nov;7(11):1944. doi: 10.1038/s41564-022-01160-6. Nat Microbiol. 2022. PMID: 36203089 Free PMC article. No abstract available.
Abstract
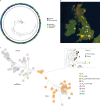
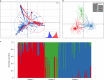
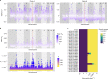
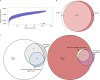
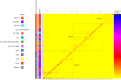
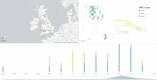
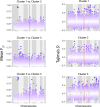
Infections caused by the fungal pathogen Aspergillus fumigatus are increasingly resistant to first-line azole antifungal drugs. However, despite its clinical importance, little is known about how susceptible patients acquire infection from drug-resistant genotypes in the environment. Here, we present a population genomic analysis of 218 A. fumigatus isolates from across the UK and Ireland (comprising 153 clinical isolates from 143 patients and 65 environmental isolates). First, phylogenomic analysis shows strong genetic structuring into two clades (A and B) with little interclade recombination and the majority of environmental azole resistance found within clade A. Second, we show occurrences where azole-resistant isolates of near-identical genotypes were obtained from both environmental and clinical sources, indicating with high confidence the infection of patients with resistant isolates transmitted from the environment. Third, genome-wide scans identified selective sweeps across multiple regions indicating a polygenic basis to the trait in some genetic backgrounds. These signatures of positive selection are seen for loci containing the canonical genes encoding fungicide resistance in the ergosterol biosynthetic pathway, while other regions under selection have no defined function. Lastly, pan-genome analysis identified genes linked to azole resistance and previously unknown resistance mechanisms. Understanding the environmental drivers and genetic basis of evolving fungal drug resistance needs urgent attention, especially in light of increasing numbers of patients with severe viral respiratory tract infections who are susceptible to opportunistic fungal superinfections.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Brown GD, et al. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4:165rv13. - PubMed
-
- Schauwvlieghe AFAD, et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir. Med. 2018;6:782–792. - PubMed
-
- Baxter CG, et al. Novel immunologic classification of aspergillosis in adult cystic fibrosis. J. Allergy Clin. Immunol. 2013;132:560–566.e10. - PubMed
-
- Kleinkauf et al. Risk Assessment on the Impact of Environmental Usage of Triazoles on the Development and Spread of Resistance to Medical Triazoles inAspergillusSpecies (ECDC, 2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

