Alfalfa (Medicago sativa L.) pho2 mutant plants hyperaccumulate phosphate
- PMID: 35471600
- PMCID: PMC9157135
- DOI: 10.1093/g3journal/jkac096
Alfalfa (Medicago sativa L.) pho2 mutant plants hyperaccumulate phosphate
Abstract
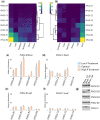
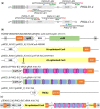
In this article, we describe a set of novel alfalfa (Medicago sativa L.) plants that hyper-accumulate Phosphate ion (Pi) at levels 3- to 6-fold higher than wild-type. This alfalfa germplasm will have practical applications reclaiming Pi from contaminated or enriched soil or be used in conservation buffer strips to protect waterways from Pi run-off. Hyper-accumulating alfalfa plants were generated by targeted mutagenesis of PHOSPHATE2 (PHO2) using newly created CRISPR/Cas9 reagents and an improved mutant screening strategy. PHO2 encodes a ubiquitin conjugating E2 enzyme (UBC24) previously characterized in Arabidopsis thaliana, Medicago truncatula, and Oryza sativa. Mutations of PHO2 disrupt Pi homeostasis resulting in Pi hyper-accumulation. Successful CRISPR/Cas9 editing of PHO2 demonstrates that this is an efficient mutagenesis tool in alfalfa despite its complex autotetraploid genome structure. Arabidopsis and M. truncatula ortholog genes were used to identify PHO2 haplotypes in outcrossing tetraploid M. sativa with the aim of generating heritable mutations in both PHO2-like genes (PHO2-B and PHO2-C). After delivery of the reagent and regeneration from transformed leaf explants, plants with mutations in all haplotypes of PHO2-B and PHO2-C were identified. These plants were evaluated for morphology, Pi accumulation, heritable transmission of targeted mutations, segregation of mutant haplotypes and removal of T-DNA(s). The Agrobacterium-mediated transformation assay and gene editing reagents reported here were also evaluated for further optimization for future alfalfa functional genomic studies.
Keywords: Agrobacterium; CRISPR/Cas9; alfalfa; haplotype; pho2.
Published by Oxford University Press on behalf of Genetics Society of America 2022. This work is written by US Government employees and is in the public domain in the US.
Figures



References
-
- Aliu E, Azanu MK, Wang and K. Lee K.. Generation of thymidine auxotrophic Agrobacterium tumefaciens strains for plant transformation. bioRxiv. 2020;1–20. DOI: 10.1101/2020.08.21.261941. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
