Single-cell RNA-Seq of human esophageal epithelium in homeostasis and allergic inflammation
- PMID: 35472002
- PMCID: PMC9208762
- DOI: 10.1172/jci.insight.159093
Single-cell RNA-Seq of human esophageal epithelium in homeostasis and allergic inflammation
Abstract
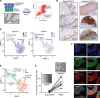
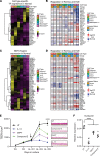
Inflammation of the esophageal epithelium is a hallmark of eosinophilic esophagitis (EoE), an emerging chronic allergic disease. Herein, we probed human esophageal epithelial cells at single-cell resolution during homeostasis and EoE. During allergic inflammation, the epithelial differentiation program was blocked, leading to loss of KRT6hi differentiated populations and expansion of TOP2hi proliferating, DSPhi transitioning, and SERPINB3hi transitioning populations; however, there was stability of the stem cell-enriched PDPNhi basal epithelial compartment. This differentiation program blockade was associated with dysregulation of transcription factors, including nuclear receptor signalers, in the most differentiated epithelial cells and altered NOTCH-related cell-to-cell communication. Each epithelial population expressed genes with allergic disease risk variants, supporting their functional interplay. The esophageal epithelium differed notably between EoE in histologic remission and controls, indicating that remission is a transitory state poised to relapse. Collectively, our data uncover the dynamic nature of the inflamed human esophageal epithelium and provide a framework to better understand esophageal health and disease.
Keywords: Cellular immune response; Immunology; Inflammation; Molecular pathology.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

