Engineering artificial photosynthetic life-forms through endosymbiosis
- PMID: 35474066
- PMCID: PMC9042829
- DOI: 10.1038/s41467-022-29961-7
Engineering artificial photosynthetic life-forms through endosymbiosis
Abstract
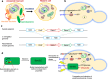
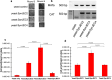
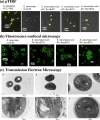
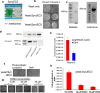
The evolutionary origin of the photosynthetic eukaryotes drastically altered the evolution of complex lifeforms and impacted global ecology. The endosymbiotic theory suggests that photosynthetic eukaryotes evolved due to endosymbiosis between non-photosynthetic eukaryotic host cells and photosynthetic cyanobacterial or algal endosymbionts. The photosynthetic endosymbionts, propagating within the cytoplasm of the host cells, evolved, and eventually transformed into chloroplasts. Despite the fundamental importance of this evolutionary event, we have minimal understanding of this remarkable evolutionary transformation. Here, we design and engineer artificial, genetically tractable, photosynthetic endosymbiosis between photosynthetic cyanobacteria and budding yeasts. We engineer various mutants of model photosynthetic cyanobacteria as endosymbionts within yeast cells where, the engineered cyanobacteria perform bioenergetic functions to support the growth of yeast cells under defined photosynthetic conditions. We anticipate that these genetically tractable endosymbiotic platforms can be used for evolutionary studies, particularly related to organelle evolution, and also for synthetic biology applications.
© 2022. The Author(s).
Conflict of interest statement
Authors declare no competing interests.
Figures

References
-
- Mereschkowsky, C. Uber natur und ursprung der chromatophoren im pflanzenreiche. Biologisches Centralblatt25, 293–604 (1905).
-
- Margulis, L. Origin of eukaryotic cells: Evidence and research implications for a theory of the origin and evolution of microbial, plant and animal cells on the precambrian Earth. (Yale University Press, 1970).
-
- Zimorski, V., Ku, C., Martin, W. F. & Gould, S. B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol.22, 38–48 (2014). - PubMed
-
- Martin, W. & Kowallik, K. V. Annotated English translation of Mereschkowsky’s 1905 paper ‘Über Natur und Ursprung der Chromatophoren im Pflanzenreiche’. Eur. J. Phycol.34, 287–295 (1999).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

