Linker histone variant HIS1-3 and WRKY1 oppositely regulate salt stress tolerance in Arabidopsis
- PMID: 35474141
- PMCID: PMC9237719
- DOI: 10.1093/plphys/kiac174
Linker histone variant HIS1-3 and WRKY1 oppositely regulate salt stress tolerance in Arabidopsis
Abstract
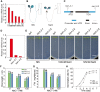
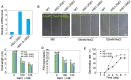
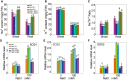
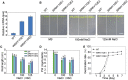
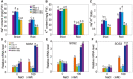
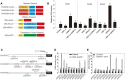
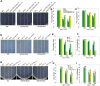
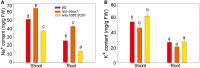
The salt overly sensitive (SOS) pathway plays an important role in plant salt stress; however, the transcriptional regulation of the genes in this pathway is unclear. In this study, we found that Linker histone variant HIS1-3 and WRKY1 oppositely regulate the salt stress response in Arabidopsis (Arabidopsis thaliana) through the transcriptional regulation of SOS genes. The expression of HIS1-3 was inhibited by salt stress, and the disruption of HIS1-3 resulted in enhanced salt tolerance. Conversely, the expression of WRKY1 was induced by salt stress, and the loss of WRKY1 function led to increased salt sensitivity. The expression of SOS1, SOS2, and SOS3 was repressed and induced by HIS1-3 and WRKY1, respectively, and HIS1-3 regulated the expression of SOS1 and SOS3 by occupying the WRKY1 binding sites on their promoters. Moreover, WRKY1 and HIS1-3 acted upstream of the SOS pathway. Together, our results indicate that HIS1-3 and WRKY1 oppositely modulate salt tolerance in Arabidopsis through transcriptional regulation of SOS genes.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
PLATZ2 negatively regulates salt tolerance in Arabidopsis seedlings by directly suppressing the expression of the CBL4/SOS3 and CBL10/SCaBP8 genes.J Exp Bot. 2020 Sep 19;71(18):5589-5602. doi: 10.1093/jxb/eraa259. J Exp Bot. 2020. PMID: 32453821
-
EIN3 and SOS2 synergistically modulate plant salt tolerance.Sci Rep. 2017 Mar 16;7:44637. doi: 10.1038/srep44637. Sci Rep. 2017. PMID: 28300216 Free PMC article.
-
CycC1;1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis.Plant Cell. 2023 Jun 26;35(7):2570-2591. doi: 10.1093/plcell/koad105. Plant Cell. 2023. PMID: 37040621 Free PMC article.
-
Revisiting plant salt tolerance: novel components of the SOS pathway.Trends Plant Sci. 2023 Sep;28(9):1060-1069. doi: 10.1016/j.tplants.2023.04.003. Epub 2023 Apr 27. Trends Plant Sci. 2023. PMID: 37117077 Review.
-
A Salt Overly Sensitive Pathway Member from Brassica juncea BjSOS3 Can Functionally Complement ΔAtsos3 in Arabidopsis.Curr Genomics. 2018 Jan;19(1):60-69. doi: 10.2174/1389202918666170228133621. Curr Genomics. 2018. PMID: 29491733 Free PMC article. Review.
Cited by
-
Molecular cloning and characterization of a salt overly sensitive3 (SOS3) gene from the halophyte Pongamia.Plant Mol Biol. 2024 May 14;114(3):57. doi: 10.1007/s11103-024-01459-4. Plant Mol Biol. 2024. PMID: 38743266
-
RcbHLH59-RcPRs module enhances salinity stress tolerance by balancing Na+/K+ through callose deposition in rose (Rosa chinensis).Hortic Res. 2022 Dec 30;10(3):uhac291. doi: 10.1093/hr/uhac291. eCollection 2023 Mar. Hortic Res. 2022. PMID: 36938564 Free PMC article.
-
Developmentally dependent reprogramming of the Arabidopsis floral transcriptome under sufficient and limited water availability.BMC Plant Biol. 2024 Apr 11;24(1):273. doi: 10.1186/s12870-024-04916-w. BMC Plant Biol. 2024. PMID: 38605371 Free PMC article.
-
Transcriptomic and Lipidomic Analysis Reveals Complex Regulation Mechanisms Underlying Rice Roots' Response to Salt Stress.Metabolites. 2024 Apr 21;14(4):244. doi: 10.3390/metabo14040244. Metabolites. 2024. PMID: 38668372 Free PMC article.
-
The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses.Cells. 2022 May 30;11(11):1796. doi: 10.3390/cells11111796. Cells. 2022. PMID: 35681491 Free PMC article.
References
-
- Agarwal P, Reddy MP, Chikara J (2011) WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep 38: 3883–3896 - PubMed
-
- Agarwal PK, Shukla PS, Gupta K, Jha B (2013) Bioengineering for salinity tolerance in plants: state of the art. Mol Biotechnol 54: 102–123 - PubMed
-
- Ascenzi R, Gantt JS (1997) A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol 34: 629–641 - PubMed
-
- Ascenzi R, Gantt JS (1999) Molecular genetic analysis of the drought-inducible linker histone variant in Arabidopsis thaliana. Plant Mol Biol 41: 159–169 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

