Unraveling the binding mode of a methamphetamine aptamer: A spectroscopic and calorimetric study
- PMID: 35474264
- PMCID: PMC9247340
- DOI: 10.1016/j.bpj.2022.04.027
Unraveling the binding mode of a methamphetamine aptamer: A spectroscopic and calorimetric study
Abstract
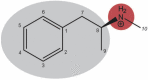
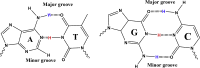
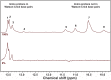
Nucleic-acid aptamers are bio-molecular recognition agents that bind to their targets with high specificity and affinity and hold promise in a range of biosensor and therapeutic applications. In the case of small-molecule targets, their small size and limited number of functional groups constitute challenges for their detection by aptamer-based biosensors because bio-recognition events may both be weak and produce poorly transduced signals. The binding affinity is principally used to characterize aptamer-ligand interactions; however, a structural understanding of bio-recognition is arguably more valuable in order to design a strong response in biosensor applications. Using a combination of nuclear magnetic resonance, circular dichroism, and isothermal titration calorimetry, we propose a binding model for a new methamphetamine aptamer and determine the main interactions driving complex formation. These measurements reveal only modest structural changes to the aptamer upon binding and are consistent with a conformational-selection binding model. The aptamer-methamphetamine complex formation was observed to be entropically driven, apparently involving hydrophobic and electrostatic interactions. Taken together, our results exemplify a means of elucidating small molecule-aptamer binding interactions, which may be decisive in the development of aptasensors and therapeutics and may contribute to a deeper understanding of interactions driving aptamer selection.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

