Two DNA vaccines protect against severe disease and pathology due to SARS-CoV-2 in Syrian hamsters
- PMID: 35474311
- PMCID: PMC9042934
- DOI: 10.1038/s41541-022-00461-5
Two DNA vaccines protect against severe disease and pathology due to SARS-CoV-2 in Syrian hamsters
Abstract
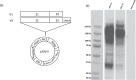
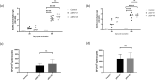
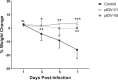
The SARS-CoV-2 pandemic is an ongoing threat to global health, and wide-scale vaccination is an efficient method to reduce morbidity and mortality. We designed and evaluated two DNA plasmid vaccines, based on the pIDV-II system, expressing the SARS-CoV-2 spike gene, with or without an immunogenic peptide, in mice, and in a Syrian hamster model of infection. Both vaccines demonstrated robust immunogenicity in BALB/c and C57BL/6 mice. Additionally, the shedding of infectious virus and the viral burden in the lungs was reduced in immunized hamsters. Moreover, high-titers of neutralizing antibodies with activity against multiple SARS-CoV-2 variants were generated in immunized animals. Vaccination also protected animals from weight loss during infection. Additionally, both vaccines were effective at reducing both pulmonary and extrapulmonary pathology in vaccinated animals. These data show the potential of a DNA vaccine for SARS-CoV-2 and suggest further investigation in large animal and human studies could be pursued.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

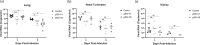
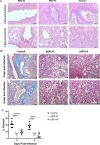
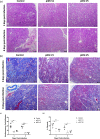

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

