Rapid synchronous type 1 IFN and virus-specific T cell responses characterize first wave non-severe SARS-CoV-2 infections
- PMID: 35474751
- PMCID: PMC8895494
- DOI: 10.1016/j.xcrm.2022.100557
Rapid synchronous type 1 IFN and virus-specific T cell responses characterize first wave non-severe SARS-CoV-2 infections
Abstract
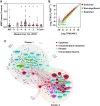
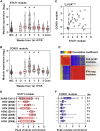
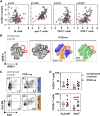
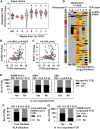
Effective control of SARS-CoV-2 infection on primary exposure may reveal correlates of protective immunity to future variants, but we lack insights into immune responses before or at the time virus is first detected. We use blood transcriptomics, multiparameter flow cytometry, and T cell receptor (TCR) sequencing spanning the time of incident non-severe infection in unvaccinated virus-naive individuals to identify rapid type 1 interferon (IFN) responses common to other acute respiratory viruses and cell proliferation responses that discriminate SARS-CoV-2 from other viruses. These peak by the time the virus is first detected and sometimes precede virus detection. Cell proliferation is most evident in CD8 T cells and associated with specific expansion of SARS-CoV-2-reactive TCRs, in contrast to virus-specific antibodies, which lag by 1-2 weeks. Our data support a protective role for early type 1 IFN and CD8 T cell responses, with implications for development of universal T cell vaccines.
Keywords: CD8 T cells; non-severe SARS-CoV-2 infection; type 1 interferon.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures

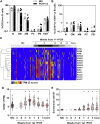
References
-
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_20060/MRC_/Medical Research Council/United Kingdom
- SP/20/2/34841/BHF_/British Heart Foundation/United Kingdom
- MRF-044-0001-RG-SWADL/MRF_/MRF_/United Kingdom
- FS/19/35/34374/BHF_/British Heart Foundation/United Kingdom
- NIHR134607/DH_/Department of Health/United Kingdom
- MR/R02622X/1/MRC_/Medical Research Council/United Kingdom
- FS/CRTF/21/24143/BHF_/British Heart Foundation/United Kingdom
- MR/N007727/1/MRC_/Medical Research Council/United Kingdom
- MR/W020610/1/MRC_/Medical Research Council/United Kingdom
- MR/S019553/1/MRC_/Medical Research Council/United Kingdom
- AA/18/6/34223/BHF_/British Heart Foundation/United Kingdom
- DRF-2018-11-ST2-004/DH_/Department of Health/United Kingdom
- 214191/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- MR/V036939/1/MRC_/Medical Research Council/United Kingdom
- 207511/Z/17/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

