Large scale genotype- and phenotype-driven machine learning in Von Hippel-Lindau disease
- PMID: 35475554
- PMCID: PMC9356987
- DOI: 10.1002/humu.24392
Large scale genotype- and phenotype-driven machine learning in Von Hippel-Lindau disease
Abstract
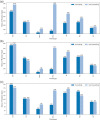
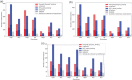
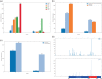
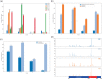
Von Hippel-Lindau (VHL) disease is a hereditary cancer syndrome where individuals are predisposed to tumor development in the brain, adrenal gland, kidney, and other organs. It is caused by pathogenic variants in the VHL tumor suppressor gene. Standardized disease information has been difficult to collect due to the rarity and diversity of VHL patients. Over 4100 unique articles published until October 2019 were screened for germline genotype-phenotype data. Patient data were translated into standardized descriptions using Human Genome Variation Society gene variant nomenclature and Human Phenotype Ontology terms and has been manually curated into an open-access knowledgebase called Clinical Interpretation of Variants in Cancer. In total, 634 unique VHL variants, 2882 patients, and 1991 families from 427 papers were captured. We identified relationship trends between phenotype and genotype data using classic statistical methods and spectral clustering unsupervised learning. Our analyses reveal earlier onset of pheochromocytoma/paraganglioma and retinal angiomas, phenotype co-occurrences and genotype-phenotype correlations including hotspots. It confirms existing VHL associations and can be used to identify new patterns and associations in VHL disease. Our database serves as an aggregate knowledge translation tool to facilitate sharing information about the pathogenicity of VHL variants.
Keywords: CIViC; Von Hippel-Lindau; genotype-phenotype; machine learning; spectral clustering.
© 2022 The Authors. Human Mutation published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
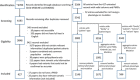
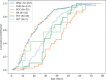
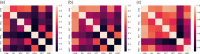
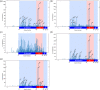
References
-
- Alosi, D. , Bisgaard, M. L. , Hemmingsen, S. N. , Krogh, L. N. , Mikkelsen, H. B. , & Binderup, M. L. M. (2017). Management of gene variants of unknown significance: Analysis method and risk assessment of the VHL mutation p.P81S (c.241C>T). Current Genomics, 18(1), 93–103. 10.2174/1389202917666160805153221 - DOI - PMC - PubMed
-
- Amendola, L. M. , Jarvik, G. P. , Leo, M. C. , McLaughlin, H. M. , Akkari, Y. , Amaral, M. D. , Berg, J. S. , Biswas, S. , Bowling, K. M. , Conlin, L. K. , Cooper, G. M. , Dorschner, M. O. , Dulik, M. C. , Ghazani, A. A. , Ghosh, R. , Green, R. C. , Hart, R. , Horton, C. , Johnston, J. J. , … Rehm, H. L. (2016). Performance of ACMG‐AMP variant‐interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. American Journal of Human Genetics, 98(6), 1067–1076. 10.1016/j.ajhg.2016.03.024 - DOI - PMC - PubMed
-
- Barlier, A. , & Mohamed, A. (2021, April 6). The UMD‐VHL mutations database. http://www.umd.be/VHL/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

