Trisomy 21 increases microtubules and disrupts centriolar satellite localization
- PMID: 35476505
- PMCID: PMC9635274
- DOI: 10.1091/mbc.E21-10-0517-T
Trisomy 21 increases microtubules and disrupts centriolar satellite localization
Abstract
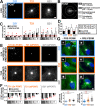
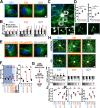
Trisomy 21, the source of Down syndrome, causes a 0.5-fold protein increase of the chromosome 21-resident gene Pericentrin (PCNT) and reduces primary cilia formation and signaling. We investigate how PCNT imbalances disrupt cilia. Using isogenic RPE-1 cells with increased chromosome 21 dosage, we find PCNT accumulates around the centrosome as a cluster of enlarged cytoplasmic puncta that localize along microtubules (MTs) and at MT ends. Cytoplasmic PCNT puncta impact the density, stability, and localization of the MT trafficking network required for primary cilia. The PCNT puncta appear to sequester cargo peripheral to centrosomes in what we call pericentrosomal crowding. The centriolar satellite proteins PCM1, CEP131, and CEP290, important for ciliogenesis, accumulate at enlarged PCNT puncta in trisomy 21 cells. Reducing PCNT when chromosome 21 ploidy is elevated is sufficient to decrease PCNT puncta and pericentrosomal crowding, reestablish a normal density of MTs around the centrosome, and restore ciliogenesis to wild-type levels. A transient reduction in MTs also decreases pericentrosomal crowding and partially rescues ciliogenesis in trisomy 21 cells, indicating that increased PCNT leads to defects in the MT network deleterious to normal centriolar satellite distribution. We propose that chromosome 21 aneuploidy disrupts MT-dependent intracellular trafficking required for primary cilia.
Figures


References
-
- Bernhard W, de Harven E (1960). L’ultrastructure du centriole et d’autres éléments de l’appareil achromatique. In Vierter Internationaler Kongress für Elektronenmikroskopie/Fourth International Conference on Electron Microscopy/Quatrième Congrès International de Microscopie Électronique: Berlin, September 10–17, 1958, Bargmann W., Möllenstedt G., Niehrs H., Peters D., Ruska E., Wolpers C. (eds.) (Springer, Berlin Heidelberg; ), pp. 217–227.
-
- Bornens M (2002). Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14, 25–34. - PubMed
-
- Bornens M, Azimzadeh J (2007). Origin and evolution of the centrosome. Adv Exp Med Biol 607, 119–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

