Genomewide CRISPR knockout screen identified PLAC8 as an essential factor for SADS-CoVs infection
- PMID: 35476513
- PMCID: PMC9170153
- DOI: 10.1073/pnas.2118126119
Genomewide CRISPR knockout screen identified PLAC8 as an essential factor for SADS-CoVs infection
Abstract
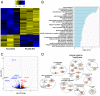
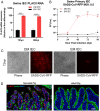
Zoonotic transmission of coronaviruses poses an ongoing threat to human populations. Endemic outbreaks of swine acute diarrhea syndrome coronavirus (SADS-CoV) have caused severe economic losses in the pig industry and have the potential to cause human outbreaks. Currently, there are no vaccines or specific antivirals against SADS-CoV, and our limited understanding of SADS-CoV host entry factors could hinder prompt responses to a potential human outbreak. Using a genomewide CRISPR knockout screen, we identified placenta-associated 8 protein (PLAC8) as an essential host factor for SADS-CoV infection. Knockout of PLAC8 abolished SADS-CoV infection, which was restored by complementing PLAC8 from multiple species, including human, rhesus macaques, mouse, pig, pangolin, and bat, suggesting a conserved infection pathway and susceptibility of SADS-CoV among mammals. Mechanistically, PLAC8 knockout does not affect viral entry; rather, knockout cells displayed a delay and reduction in viral subgenomic RNA expression. In a swine primary intestinal epithelial culture (IEC) infection model, differentiated cultures have high levels of PLAC8 expression and support SADS-CoV replication. In contrast, expanding IECs have low levels of PLAC8 expression and are resistant to SADS-CoV infection. PLAC8 expression patterns translate in vivo; the immunohistochemistry of swine ileal tissue revealed high levels of PLAC8 protein in neonatal compared to adult tissue, mirroring the known SADS-CoV pathogenesis in neonatal piglets. Overall, PLAC8 is an essential factor for SADS-CoV infection and may serve as a promising target for antiviral development for potential pandemic SADS-CoV.
Keywords: CRISPR; PLAC8; coronavirus; swine acute diarrhea syndrome coronavirus.
Conflict of interest statement
The authors declare no competing interest.
Figures






References
-
- Drosten C., et al. , Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003). - PubMed
-
- Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D. M. E., Fouchier R. A. M., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

