Distinguishing Admissions Specifically for COVID-19 From Incidental SARS-CoV-2 Admissions: National Retrospective Electronic Health Record Study
- PMID: 35476727
- PMCID: PMC9119395
- DOI: 10.2196/37931
Distinguishing Admissions Specifically for COVID-19 From Incidental SARS-CoV-2 Admissions: National Retrospective Electronic Health Record Study
Abstract
Background: Admissions are generally classified as COVID-19 hospitalizations if the patient has a positive SARS-CoV-2 polymerase chain reaction (PCR) test. However, because 35% of SARS-CoV-2 infections are asymptomatic, patients admitted for unrelated indications with an incidentally positive test could be misclassified as a COVID-19 hospitalization. Electronic health record (EHR)-based studies have been unable to distinguish between a hospitalization specifically for COVID-19 versus an incidental SARS-CoV-2 hospitalization. Although the need to improve classification of COVID-19 versus incidental SARS-CoV-2 is well understood, the magnitude of the problems has only been characterized in small, single-center studies. Furthermore, there have been no peer-reviewed studies evaluating methods for improving classification.
Objective: The aims of this study are to, first, quantify the frequency of incidental hospitalizations over the first 15 months of the pandemic in multiple hospital systems in the United States and, second, to apply electronic phenotyping techniques to automatically improve COVID-19 hospitalization classification.
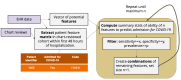
Methods: From a retrospective EHR-based cohort in 4 US health care systems in Massachusetts, Pennsylvania, and Illinois, a random sample of 1123 SARS-CoV-2 PCR-positive patients hospitalized from March 2020 to August 2021 was manually chart-reviewed and classified as "admitted with COVID-19" (incidental) versus specifically admitted for COVID-19 ("for COVID-19"). EHR-based phenotyping was used to find feature sets to filter out incidental admissions.
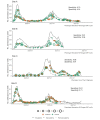
Results: EHR-based phenotyped feature sets filtered out incidental admissions, which occurred in an average of 26% of hospitalizations (although this varied widely over time, from 0% to 75%). The top site-specific feature sets had 79%-99% specificity with 62%-75% sensitivity, while the best-performing across-site feature sets had 71%-94% specificity with 69%-81% sensitivity.
Conclusions: A large proportion of SARS-CoV-2 PCR-positive admissions were incidental. Straightforward EHR-based phenotypes differentiated admissions, which is important to assure accurate public health reporting and research.
Keywords: COVID-19; SARS-CoV-2; clinical research informatics; electronic health records; health care; health data; medical informatics; patient data; phenotype; public health.
©Jeffrey G Klann, Zachary H Strasser, Meghan R Hutch, Chris J Kennedy, Jayson S Marwaha, Michele Morris, Malarkodi Jebathilagam Samayamuthu, Ashley C Pfaff, Hossein Estiri, Andrew M South, Griffin M Weber, William Yuan, Paul Avillach, Kavishwar B Wagholikar, Yuan Luo, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), Gilbert S Omenn, Shyam Visweswaran, John H Holmes, Zongqi Xia, Gabriel A Brat, Shawn N Murphy. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 18.05.2022.
Conflict of interest statement
Conflicts of Interest: The authors declare that they have no conflicts of interest. JGK reports a consulting relationship with the i2b2-tranSMART Foundation through Invocate, Inc. CJK reports consulting for the University of California, Berkeley; the University of Southern California (USC), and the University of California, San Francisco (UCSF). AMS reports funding from the National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK127208, NIH/National Heart, Lung, and Blood Institute (NHLBI) R01HL146818, and institutional pilot awards from the Wake Forest School of Medicine. GMW reports consulting for the i2b2-tranSMART Foundation. PA reports consulting for the Cincinnati Children’s Hospital Medical Center (CCHMC) and Boston Children’s Hospital (BCH). ZX has received research support from the NIH, the Department of Defense, and Octave Biosciences and has served on the scientific advisory board for Genentech/Roche. SNM reports professional relationships with the Scientific Advisory Board for Boston University, the Universidad de Puerto Rico, the University of California, Los Angeles (UCLA), the University of Massachusetts Medical School (UMMS), and the Kenner Family Research Fund.
Figures

Update of
-
Distinguishing Admissions Specifically for COVID-19 from Incidental SARS-CoV-2 Admissions: A National EHR Research Consortium Study.medRxiv [Preprint]. 2022 Feb 18:2022.02.10.22270728. doi: 10.1101/2022.02.10.22270728. medRxiv. 2022. Update in: J Med Internet Res. 2022 May 18;24(5):e37931. doi: 10.2196/37931. PMID: 35350202 Free PMC article. Updated. Preprint.
References
-
- Haendel MA, Chute CG, Bennett TD, Eichmann DA, Guinney J, Kibbe WA, Payne PRO, Pfaff ER, Robinson PN, Saltz JH, Spratt H, Suver C, Wilbanks J, Wilcox AB, Williams AE, Wu C, Blacketer C, Bradford RL, Cimino JJ, Clark M, Colmenares EW, Francis PA, Gabriel D, Graves A, Hemadri R, Hong SS, Hripscak G, Jiao D, Klann JG, Kostka K, Lee AM, Lehmann HP, Lingrey L, Miller RT, Morris M, Murphy SN, Natarajan K, Palchuk MB, Sheikh U, Solbrig H, Visweswaran S, Walden A, Walters KM, Weber GM, Zhang XT, Zhu RL, Amor B, Girvin AT, Manna A, Qureshi N, Kurilla MG, Michael SG, Portilla LM, Rutter JL, Austin CP, Gersing KR, N3C Consortium The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021 Mar 01;28(3):427–443. doi: 10.1093/jamia/ocaa196. http://europepmc.org/abstract/MED/32805036 5893482 - DOI - PMC - PubMed
-
- Brat GA, Weber GM, Gehlenborg N, Avillach P, Palmer NP, Chiovato L, Cimino J, Waitman LR, Omenn GS, Malovini A, Moore JH, Beaulieu-Jones BK, Tibollo V, Murphy SN, Yi SL, Keller MS, Bellazzi R, Hanauer DA, Serret-Larmande A, Gutierrez-Sacristan A, Holmes JJ, Bell DS, Mandl KD, Follett RW, Klann JG, Murad DA, Scudeller L, Bucalo M, Kirchoff K, Craig J, Obeid J, Jouhet V, Griffier R, Cossin S, Moal B, Patel LP, Bellasi A, Prokosch HU, Kraska D, Sliz P, Tan ALM, Ngiam KY, Zambelli A, Mowery DL, Schiver E, Devkota B, Bradford RL, Daniar M, Daniel C, Benoit V, Bey R, Paris N, Serre P, Orlova N, Dubiel J, Hilka M, Jannot AS, Breant S, Leblanc J, Griffon N, Burgun A, Bernaux M, Sandrin A, Salamanca E, Cormont S, Ganslandt T, Gradinger T, Champ J, Boeker M, Martel P, Esteve L, Gramfort A, Grisel O, Leprovost D, Moreau T, Varoquaux G, Vie J, Wassermann D, Mensch A, Caucheteux C, Haverkamp C, Lemaitre G, Bosari S, Krantz ID, South A, Cai T, Kohane IS. International electronic health record-derived COVID-19 clinical course profiles: the 4CE consortium. NPJ Digit Med. 2020;3:109. doi: 10.1038/s41746-020-00308-0. doi: 10.1038/s41746-020-00308-0.308 - DOI - DOI - PMC - PubMed
-
- Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(2):124–30. doi: 10.1136/jamia.2009.000893. http://europepmc.org/abstract/MED/20190053 17/2/124 - DOI - PMC - PubMed
-
- Visweswaran S, Samayamuthu MJ, Morris M, Weber GM, MacFadden D, Trevvett P, Klann JG, Gainer VS, Benoit B, Murphy SN, Patel L, Mirkovic N, Borovskiy Y, Johnson RD, Wyatt MC, Wang AY, Follett RW, Chau N, Zhu W, Abajian M, Chuang A, Bahroos N, Reeder P, Xie D, Cai J, Sendro ER, Toto RD, Firestein GS, Nadler LM, Reis SE. Development of a coronavirus disease 2019 (COVID-19) application ontology for the Accrual to Clinical Trials (ACT) network. JAMIA Open. 2021 Apr;4(2):ooab036. doi: 10.1093/jamiaopen/ooab036. http://europepmc.org/abstract/MED/34113801 ooab036 - DOI - PMC - PubMed
-
- Khullar D. Do the Omicron Numbers Mean What We Think They Mean? The New Yorker. 2022. Jan 16, [2022-01-25]. https://www.newyorker.com/magazine/2022/01/24/do-the-omicron-numbers-mea... .
Publication types
MeSH terms
Grants and funding
- UL1 TR000005/TR/NCATS NIH HHS/United States
- UL1 TR001878/TR/NCATS NIH HHS/United States
- R01 HG009174/HG/NHGRI NIH HHS/United States
- R01 NS098023/NS/NINDS NIH HHS/United States
- R01 NS124882/NS/NINDS NIH HHS/United States
- R01 HL151643/HL/NHLBI NIH HHS/United States
- U01 TR003528/TR/NCATS NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- P30 ES017885/ES/NIEHS NIH HHS/United States
- UL1 TR001857/TR/NCATS NIH HHS/United States
- T15 LM007092/LM/NLM NIH HHS/United States
- R01 LM013345/LM/NLM NIH HHS/United States
- UL1 TR002541/TR/NCATS NIH HHS/United States
- FS/19/52/34563/BHF_/British Heart Foundation/United Kingdom
- T32 HD040128/HD/NICHD NIH HHS/United States
- R01 LM013337/LM/NLM NIH HHS/United States
- K23 HL148394/HL/NHLBI NIH HHS/United States
- R01 HL146818/HL/NHLBI NIH HHS/United States
- L40 HL148910/HL/NHLBI NIH HHS/United States
- R01 DK127208/DK/NIDDK NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

