In vivo partial cellular reprogramming enhances liver plasticity and regeneration
- PMID: 35476977
- PMCID: PMC9807246
- DOI: 10.1016/j.celrep.2022.110730
In vivo partial cellular reprogramming enhances liver plasticity and regeneration
Abstract
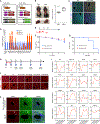
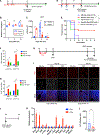
Mammals have limited regenerative capacity, whereas some vertebrates, like fish and salamanders, are able to regenerate their organs efficiently. The regeneration in these species depends on cell dedifferentiation followed by proliferation. We generate a mouse model that enables the inducible expression of the four Yamanaka factors (Oct-3/4, Sox2, Klf4, and c-Myc, or 4F) specifically in hepatocytes. Transient in vivo 4F expression induces partial reprogramming of adult hepatocytes to a progenitor state and concomitantly increases cell proliferation. This is indicated by reduced expression of differentiated hepatic-lineage markers, an increase in markers of proliferation and chromatin modifiers, global changes in DNA accessibility, and an acquisition of liver stem and progenitor cell markers. Functionally, short-term expression of 4F enhances liver regenerative capacity through topoisomerase2-mediated partial reprogramming. Our results reveal that liver-specific 4F expression in vivo induces cellular plasticity and counteracts liver failure, suggesting that partial reprogramming may represent an avenue for enhancing tissue regeneration.
Keywords: CP: Stem cell research; dedifferentiation; liver; regeneration; reprogramming.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Abad M, Mosteiro L, Pantoja C, Canamero M, Rayon T, Ors I, Grana O, Megias D, Dominguez O, Martinez D, et al. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340–345. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

