Microbial Dark Matter: from Discovery to Applications
- PMID: 35477055
- PMCID: PMC10025686
- DOI: 10.1016/j.gpb.2022.02.007
Microbial Dark Matter: from Discovery to Applications
Abstract
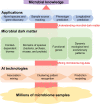
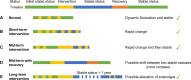
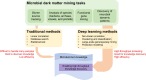
With the rapid increase of the microbiome samples and sequencing data, more and more knowledge about microbial communities has been gained. However, there is still much more to learn about microbial communities, including billions of novel species and genes, as well as countless spatiotemporal dynamic patterns within the microbial communities, which together form the microbial dark matter. In this work, we summarized the dark matter in microbiome research and reviewed current data mining methods, especially artificial intelligence (AI) methods, for different types of knowledge discovery from microbial dark matter. We also provided case studies on using AI methods for microbiome data mining and knowledge discovery. In summary, we view microbial dark matter not as a problem to be solved but as an opportunity for AI methods to explore, with the goal of advancing our understanding of microbial communities, as well as developing better solutions to global concerns about human health and the environment.
Keywords: Application; Artificial intelligence; Dark matter; Knowledge discovery; Microbiome.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., et al. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

