Functional interaction between compound heterozygous TERT mutations causes severe telomere biology disorder
- PMID: 35477117
- PMCID: PMC9631560
- DOI: 10.1182/bloodadvances.2022007029
Functional interaction between compound heterozygous TERT mutations causes severe telomere biology disorder
Abstract
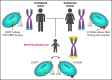
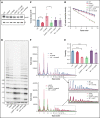
Telomere biology disorders (TBDs) are a spectrum of multisystem inherited disorders characterized by bone marrow failure, resulting from mutations in the genes encoding telomerase or other proteins involved in maintaining telomere length and integrity. Pathogenicity of variants in these genes can be hard to evaluate, because TBD mutations show highly variable penetrance and genetic anticipation related to inheritance of shorter telomeres with each generation. Thus, detailed functional analysis of newly identified variants is often essential. Herein, we describe a patient with compound heterozygous variants in the TERT gene, which encodes the catalytic subunit of telomerase, hTERT. This patient had the extremely severe Hoyeraal-Hreidarsson form of TBD, although his heterozygous parents were clinically unaffected. Molecular dynamic modeling and detailed biochemical analyses demonstrate that one allele (L557P) affects association of hTERT with its cognate RNA component hTR, whereas the other (K1050E) affects the binding of telomerase to its DNA substrate and enzyme processivity. Unexpectedly, the data demonstrate a functional interaction between the proteins encoded by the two alleles, with wild-type hTERT rescuing the effect of K1050E on processivity, whereas L557P hTERT does not. These data contribute to the mechanistic understanding of telomerase, indicating that RNA binding in one hTERT molecule affects the processivity of telomere addition by the other molecule. This work emphasizes the importance of functional characterization of TERT variants to reach a definitive molecular diagnosis for patients with TBD, and, in particular, it illustrates the importance of analyzing the effects of compound heterozygous variants in combination, to reveal interallelic effects.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures



References
-
- Engelhardt M, Kumar R, Albanell J, Pettengell R, Han W, Moore MA. Telomerase regulation, cell cycle, and telomere stability in primitive hematopoietic cells. Blood. 1997;90(1):182-193. - PubMed
-
- Barbaro PM, Ziegler DS, Reddel RR. The wide-ranging clinical implications of the short telomere syndromes. Intern Med J. 2016;46(4):393-403. - PubMed

