Proton pump inhibitors induce changes in the gut microbiome composition of systemic lupus erythematosus patients
- PMID: 35477382
- PMCID: PMC9043501
- DOI: 10.1186/s12866-022-02533-x
Proton pump inhibitors induce changes in the gut microbiome composition of systemic lupus erythematosus patients
Abstract
Background: Currently, few studies focus on the association between gut microbiota and systemic lupus erythematosus (SLE), and much less studies consider the effect of drug usage. Proton pump inhibitors (PPIs) are commonly used to treat drug-related gastrointestinal damage in SLE patients. Therefore, the purpose of this study is to examine the gut microbiota of SLE patients using PPIs.
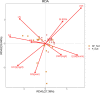
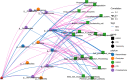
Methods: Fecal samples from 20 SLE patients with PPIs (P-SLE), 20 SLE patients without PPIs (NP-SLE) and 17 healthy controls (HCs) were obtained. The structure of the bacterial community in the fecal samples was analyzed by 16S rRNA gene sequencing. Redundancy analysis (RDA) was performed to observe the relationship between clinical variables and microbiome composition in P-SLE and NP-SLE patients. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, functional capabilities of microbiota were estimated. Network analysis was performed to analyze the association of metabolic pathway alterations with altered gut microbiota in P-SLE and NP-SLE patients.
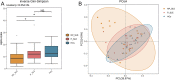
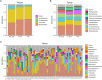
Results: P-SLE patients exhibited increased alpha-diversity and an altered composition of the gut microbiota compared with NP-SLE patients. The alpha-diversity of NP-SLE patients was significantly lower than HCs but also of P-SLE patients, whose alpha-diversity had become similar to HCs. Compared with NP-SLE patients, the relative abundances of Lactobacillus, Roseburia, Oxalobacter, and Desulfovibrio were increased, while those of Veillonella, Escherichia, Morganella, Pseudomonas and Stenotrophomonas were decreased in P-SLE patients. RDA indicated that PPI use was the only significant exploratory variable for the microbiome composition when comparing SLE patients. KEGG analysis showed that 16 metabolic pathways were significantly different between NP-SLE and P-SLE patients. These metabolic pathways were mainly associated with changes in Escherichia, Roseburia, Stenotrophomonas, Morganella and Alipipes as determined by the network analysis.
Conclusions: PPI use is associated with an improved microbiome composition of SLE patients as it 1) increases alpha-diversity levels back to normal, 2) increases the abundance of various (beneficial) commensals, and 3) decreases the abundance of certain opportunistic pathogenic genera such as Escherichia. Validation studies with higher patient numbers are however recommended to explore these patterns in more detail.
Keywords: Gut microbiota; Proton pump inhibitors; Systemic lupus erythematosus.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest concerning this article.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

