Activation of innate-adaptive immune machinery by poly(I:C) exposes a therapeutic vulnerability to prevent relapse in stroma-rich colon cancer
- PMID: 35477539
- PMCID: PMC9664095
- DOI: 10.1136/gutjnl-2021-326183
Activation of innate-adaptive immune machinery by poly(I:C) exposes a therapeutic vulnerability to prevent relapse in stroma-rich colon cancer
Abstract
Objective: Stroma-rich tumours represent a poor prognostic subtype in stage II/III colon cancer (CC), with high relapse rates and limited response to standard adjuvant chemotherapy.
Design: To address the lack of efficacious therapeutic options for patients with stroma-rich CC, we stratified our human tumour cohorts according to stromal content, enabling identification of the biology underpinning relapse and potential therapeutic vulnerabilities specifically within stroma-rich tumours that could be exploited clinically. Following human tumour-based discovery and independent clinical validation, we use a series of in vitro and stroma-rich in vivo models to test and validate the therapeutic potential of elevating the biology associated with reduced relapse in human tumours.
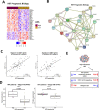
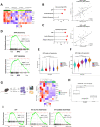
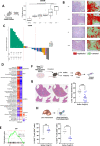
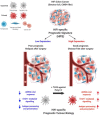
Results: By performing our analyses specifically within the stroma-rich/high-fibroblast (HiFi) subtype of CC, we identify and validate the clinical value of a HiFi-specific prognostic signature (HPS), which stratifies tumours based on STAT1-related signalling (High-HPS v Low-HPS=HR 0.093, CI 0.019 to 0.466). Using in silico, in vitro and in vivo models, we demonstrate that the HPS is associated with antigen processing and presentation within discrete immune lineages in stroma-rich CC, downstream of double-stranded RNA and viral response signalling. Treatment with the TLR3 agonist poly(I:C) elevated the HPS signalling and antigen processing phenotype across in vitro and in vivo models. In an in vivo model of stroma-rich CC, poly(I:C) treatment significantly increased systemic cytotoxic T cell activity (p<0.05) and reduced liver metastases (p<0.0002).
Conclusion: This study reveals new biological insight that offers a novel therapeutic option to reduce relapse rates in patients with the worst prognosis CC.
Keywords: ADJUVANT TREATMENT; CANCER; COLON CARCINOGENESIS; COLORECTAL CANCER.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: ML has received honoraria from Pfizer, EMF Serono and Roche for presentations unrelated to this work; ML has received an unrestricted educational grant from Pfizer for research unrelated to this work. PT has received honoraria and travel expenses from BMS, Merck, Roche, Lilly and Sanofi-Aventis for contributions that are not related to the present work. The authors declare no other potential conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- A28223/CRUK_/Cancer Research UK/United Kingdom
- 24387/CRUK_/Cancer Research UK/United Kingdom
- 110371/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_17117/MRC_/Medical Research Council/United Kingdom
- A26825/CRUK_/Cancer Research UK/United Kingdom
- MR/S021205/1/MRC_/Medical Research Council/United Kingdom
- G0400302/MRC_/Medical Research Council/United Kingdom
- MR/M016587/1/MRC_/Medical Research Council/United Kingdom
- 29834/CRUK_/Cancer Research UK/United Kingdom
- A21139/CRUK_/Cancer Research UK/United Kingdom
- 26045/CRUK_/Cancer Research UK/United Kingdom
- A17196/CRUK_/Cancer Research UK/United Kingdom
- A31287/CRUK_/Cancer Research UK/United Kingdom
- 15333/CRUK_/Cancer Research UK/United Kingdom
- BB/T002824/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 206314/Z/17/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
