Molecular Characterization of Membranous Nephropathy
- PMID: 35477557
- PMCID: PMC9161788
- DOI: 10.1681/ASN.2021060784
Molecular Characterization of Membranous Nephropathy
Abstract
Background: Molecular characterization of nephropathies may facilitate pathophysiologic insight, development of targeted therapeutics, and transcriptome-based disease classification. Although membranous nephropathy (MN) is a common cause of adult-onset nephrotic syndrome, the molecular pathways of kidney damage in MN require further definition.
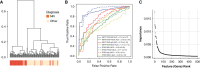
Methods: We applied a machine-learning framework to predict diagnosis on the basis of gene expression from the microdissected kidney tissue of participants in the Nephrotic Syndrome Study Network (NEPTUNE) cohort. We sought to identify differentially expressed genes between participants with MN versus those of other glomerulonephropathies across the NEPTUNE and European Renal cDNA Bank (ERCB) cohorts, to find MN-specific gene modules in a kidney-specific functional network, and to identify cell-type specificity of MN-specific genes using single-cell sequencing data from reference nephrectomy tissue.
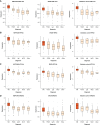
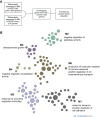
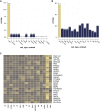
Results: Glomerular gene expression alone accurately separated participants with MN from those with other nephrotic syndrome etiologies. The top predictive classifier genes from NEPTUNE participants were also differentially expressed in the ERCB participants with MN. We identified a signature of 158 genes that are significantly differentially expressed in MN across both cohorts, finding 120 of these in a validation cohort. This signature is enriched in targets of transcription factor NF-κB. Clustering these MN-specific genes in a kidney-specific functional network uncovered modules with functional enrichments, including in ion transport, cell projection morphogenesis, regulation of adhesion, and wounding response. Expression data from reference nephrectomy tissue indicated 43% of these genes are most highly expressed by podocytes.
Conclusions: These results suggest that, relative to other glomerulonephropathies, MN has a distinctive molecular signature that includes upregulation of many podocyte-expressed genes, provides a molecular snapshot of MN, and facilitates insight into MN's underlying pathophysiology.
Keywords: machine learning; membranous nephropathy; podocyte; scRNA-seq; single-cell sequencing; transcriptional profiling.
Copyright © 2022 by the American Society of Nephrology.
Figures


Comment in
-
Molecular Characterization of Membranous Nephropathy: Quo Vadis?.J Am Soc Nephrol. 2022 Jun;33(6):1057-1059. doi: 10.1681/ASN.2022040395. Epub 2022 May 16. J Am Soc Nephrol. 2022. PMID: 35577559 Free PMC article. No abstract available.
-
In Silico-Based Approach to the Discovery of New Antigens in Membranous Nephropathy.J Am Soc Nephrol. 2022 Dec;33(12):2321-2322. doi: 10.1681/ASN.2022070832. Epub 2022 Sep 29. J Am Soc Nephrol. 2022. PMID: 36175143 Free PMC article. No abstract available.
-
Authors' Reply: In Silico-Based Approach to the Discovery of New Antigens in Membranous Nephropathy.J Am Soc Nephrol. 2022 Dec;33(12):2322-2323. doi: 10.1681/ASN.2022080921. Epub 2022 Sep 29. J Am Soc Nephrol. 2022. PMID: 36175144 Free PMC article. No abstract available.
References
-
- Sethi S, Debiec H, Madden B, Charlesworth MC, Morelle J, Gross L, et al. : Neural epidermal growth factor-like 1 protein (NELL-1) associated membranous nephropathy. Kidney Int 97: 163–174, 2020 - PubMed
-
- Bobart SA, Tehranian S, Sethi S, Alexander MP, Nasr SH, Moura Marta C, et al. : A target antigen-based approach to the classification of membranous nephropathy. Mayo Clin Proc 96: 577–591, 2021 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

