Comprehensive human amniotic fluid metagenomics supports the sterile womb hypothesis
- PMID: 35477737
- PMCID: PMC9046152
- DOI: 10.1038/s41598-022-10869-7
Comprehensive human amniotic fluid metagenomics supports the sterile womb hypothesis
Abstract
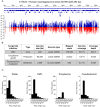
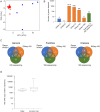
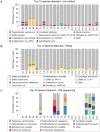
As metagenomic approaches for detecting infectious agents have improved, each tissue that was once thought to be sterile has been found to harbor a variety of microorganisms. Controversy still exists over the status of amniotic fluid, which is part of an immunologically privileged zone that is required to prevent maternal immune system rejection of the fetus. Due to this privilege, the exclusion of microbes has been proposed to be mandatory, leading to the sterile womb hypothesis. Since nucleic acid yields from amniotic fluid are very low, contaminating nucleic acid found in water, reagents and the laboratory environment frequently confound attempts to address this hypothesis. Here we present metagenomic criteria for microorganism detection and a metagenomic method able to be performed with small volumes of starting material, while controlling for exogenous contamination, to circumvent these and other pitfalls. We use this method to show that human mid-gestational amniotic fluid has no detectable virome or microbiome, supporting the sterile womb hypothesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Remington, J. S., Klein, J. O., Nizet, V., Maldonado, Y. & Wilson, C. B. Infectious Diseases of the Fetus and Newborn Infant. (Saunders/Elsevier, 2011).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

