TLR7 gain-of-function genetic variation causes human lupus
- PMID: 35477763
- PMCID: PMC9095492
- DOI: 10.1038/s41586-022-04642-z
TLR7 gain-of-function genetic variation causes human lupus
Abstract
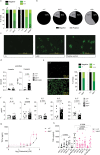
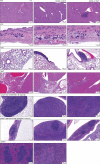
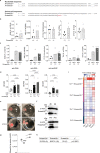
Although circumstantial evidence supports enhanced Toll-like receptor 7 (TLR7) signalling as a mechanism of human systemic autoimmune disease1-7, evidence of lupus-causing TLR7 gene variants is lacking. Here we describe human systemic lupus erythematosus caused by a TLR7 gain-of-function variant. TLR7 is a sensor of viral RNA8,9 and binds to guanosine10-12. We identified a de novo, previously undescribed missense TLR7Y264H variant in a child with severe lupus and additional variants in other patients with lupus. The TLR7Y264H variant selectively increased sensing of guanosine and 2',3'-cGMP10-12, and was sufficient to cause lupus when introduced into mice. We show that enhanced TLR7 signalling drives aberrant survival of B cell receptor (BCR)-activated B cells, and in a cell-intrinsic manner, accumulation of CD11c+ age-associated B cells and germinal centre B cells. Follicular and extrafollicular helper T cells were also increased but these phenotypes were cell-extrinsic. Deficiency of MyD88 (an adaptor protein downstream of TLR7) rescued autoimmunity, aberrant B cell survival, and all cellular and serological phenotypes. Despite prominent spontaneous germinal-centre formation in Tlr7Y264H mice, autoimmunity was not ameliorated by germinal-centre deficiency, suggesting an extrafollicular origin of pathogenic B cells. We establish the importance of TLR7 and guanosine-containing self-ligands for human lupus pathogenesis, which paves the way for therapeutic TLR7 or MyD88 inhibition.
© 2022. The Author(s).
Conflict of interest statement
M.P.G. is working with Pharmorage Pty on the therapeutic development of TLR7 inhibitors. The other authors declare no competing interests.
Figures










Comment in
-
Novel TLR7 variant causes lupus.Nat Rev Rheumatol. 2022 Jul;18(7):365. doi: 10.1038/s41584-022-00796-2. Nat Rev Rheumatol. 2022. PMID: 35610363 No abstract available.
-
Resolving a paradox between mouse and man: a genetic link between TLR7 and the pathogenesis of human lupus nephritis.Kidney Int. 2023 May;103(5):824-826. doi: 10.1016/j.kint.2023.01.019. Kidney Int. 2023. PMID: 37085253 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

